-
PDF
- Split View
-
Views
-
Cite
Cite
Fernanda G. Paião, Fernando Segato, Jeny R. Cursino-Santos, Nalu T.A. Peres, Nilce M. Martinez-Rossi, Analysis of Trichophyton rubrum gene expression in response to cytotoxic drugs, FEMS Microbiology Letters, Volume 271, Issue 2, June 2007, Pages 180–186, https://doi.org/10.1111/j.1574-6968.2007.00710.x
Close - Share Icon Share
Abstract
Suppressive subtractive hybridization was used to isolate transcripts specifically upregulated during Trichophyton rubrum exposure to acriflavin, fluconazole, griseofulvin, terbinafine or undecanoic acid. Macro-array dot-blot and sequencing of 132 clones, which correspond to genes differentially expressed after exposition of T. rubrum to at least one of these cytotoxic drugs, revealed 39 unique genes. Of these, 32 have not been previously described in T. rubrum, representing an increase in the number of T. rubrum genes that have been identified. The upregulation of the novel genes encoding a retrotransposon element, a carboxylic ester hydrolase, a copper resistance-associated P-type ATPase, a DNA mismatch repair protein and a NIMA (never in mitosis A) interactive protein was confirmed by Northern blot.
Introduction
Trichophyton rubrum is a dermatophyte that represents the most prevalent fungus isolated from skin and nail lesions of humans (Baudraz-Rosselet et al., 2005). Currently available antifungal agents against dermatophytes are few and have limitations regarding efficacy and toxicity. Furthermore, the improper use of antimycotics, the practice of invasive medical procedures and the onset of AIDS have led to cases of reincidence of mycoses caused by T. rubrum. In addition, deep infections caused by this pathogen have been reported in immunocompromised hosts suggesting that new therapeutic strategies are necessary (Nir-Paz et al., 2003; Cordeiro et al., 2006). In this way, the aim of this study was to investigate changes in the gene expression profile of T. rubrum following exposure to acriflavin, fluconazole, griseofulvin, terbinafine or undecanoic acid, which represent different classes of antifungal agents.
Terbinafine interferes with ergosterol biosynthesis, the major membrane sterol of fungi, by inhibiting squalene epoxidase, resulting in ergosterol deficiency in the fungal membrane and, thus, inhibition of fungal growth (Ghannoum et al., 1994, 2004). Griseofulvin interferes with the function of spindle microtubules, inhibiting nucleic acid synthesis and cell mitosis. It is also an antagonist of chitin synthesis in the fungal cell wall (Georgopapadakou & Walsh, 1996). Fluconazole, a fungistatic agent, inhibits the cytochrome 450 sterol 14 α-demethylase, affecting the conversion of lanosterol into ergosterol, leading to disruption of fungal membranes, accumulation of phospholipids within the cell and cell death (Pasko et al., 1990; Sheehan et al., 1999). Other cytotoxic agents used in this work are acriflavin, which interferes with replication, transcription and translation processes by intercalating RNA and DNA molecules, and undecanoic acid, a fatty acid with antimycotic properties that inhibits the production of exocellular keratinase, lipase and the biosynthesis of several phospholipids in T. rubrum (Das & Banerjee, 1982a, b).
Here, the construction of a subtractive suppressive cDNA library and the isolation of transcripts specifically expressed during T. rubrum exposure to acriflavin, fluconazole, griseofulvin, terbinafine or undecanoic acid are reported. This subtractive cDNA library allows us to carry out functional analysis of these genes to better understand the mechanisms of action of these drugs and the mechanism of stress adaptation. It will also help in the search for new targets to antifungal drugs.
Materials and methods
Fungal strain and growth conditions
The clinical isolate H6 (ATCC MYA-3108) of T. rubrum used throughout this study was cultured as previously described (Fachin et al., 1996). For gene expression assay, T. rubrum mycelia obtained from Sabouraud agar-plates cultured for 10–15 days were inoculated in liquid Sabouraud and incubated at 28°C for 72 h on an orbital shaker at 180 r.p.m. The resulting mycelia were aseptically transferred to fresh Sabouraud medium, to which was added acriflavin (2.5 µg mL−1), fluconazole (300 µg mL−1), griseofulvin (2.0 µg mL−1), terbinafine (0.1 µg mL−1) or undecanoic acid (50 µg mL−1), which correspond to 90% of their minimal inhibitory concentrations (MICs) (Cervelatti et al., 2006). As a control, the mycelia were inoculated into drug-free medium. The cultures were incubated for 15 min under the same conditions mentioned above, harvested by filtration and used for RNA extraction.
RNA isolation and construction of subtractive cDNA library
Total RNA was extracted from about 100 mg frozen grounded mycelium using the Purescript RNA isolation Kit (Gentra Systems). Poly (A+) was isolated from total RNA using Oligotex mRNA spin columns following the manufacturer's procedure (Quiagen).
Suppression subtractive hybridization (SSH) was performed between the tester (mixture of mRNAs from H6 mycelia exposed to each cytotoxic compound) and driver (mRNA obtained from H6 mycelia inoculated into drug-free medium). The library construction was performed as previously described (Diatchenko et al., 1996), using the Clontech PCR-Select cDNA Subtraction Kit (BD Biosciences, Palo Alto, CA). The subtracted target cDNAs were cloned into pCR 2.1 (TOPO TA Cloning system, Invitrogen) and transformed into Escherichia coli Mos-Blue-competent cells.
DNA sequencing and analysis
The recombinant cDNA clones were sequenced on an ABI Prism® 377 DNA sequencer (Applied Biosystems), using M13 forward and reverse primers. The sequences were processed using the phredphrap-consed package, and subjected to similarity searches against the nonredundant GenBank database using blastx and the GenBank est database using blastn (Altschul et al., 1997). A match was considered significant if the expected (E) value was <10e−3. The sequences were categorized into Munich Information Center for Protein Sequences (MIPS) functional categories according to their putative blastx identification (Mewes et al., 1997, 2002). Expressed sequence tags (ESTs) were submitted to the GenBank est database under the accession numbers given in Table 1.
Expression and homologous sequences of putative genes from Trichophyton rubrum SSH library
| Change in fold expression | |||||||||
| Clones(GenBank Accession No.) | Size (bp) | Putative ID | E-value | ACR | FLC | GRS | TRB | UDA | MIPs functional category |
| TR0001 (DW005348) | 578 | Aspergillus nidulans Hypothetical protein XP_663997 | 8e−05 | 3.1 | 6.0 | 3.5 | 10.0 | 4.3 | 99 |
| TR0002 (DW005349) | 415 | Glomerella cingulata Pol protein AF264028 | 1e−42 | 3.1 | 10.9 | 6.0 | 2.9 | 1.1 | 38 |
| TR0003 (DW005350) | 156 | Aspergillus fumigatus DEAD helicases superfamily EAL84582 | 8e−20 | 7.9 | 13.4 | 6.0 | 7.9 | 4.8 | 11 |
| TR0004 (DW005351) | 229 | A. fumigatus Kynurenine aminotransferase XP_751710 | 3e−28 | 0.6 | 6.3 | 1.2 | 4.0 | 1.9 | 01 |
| TR0005 (DW005352) | 197 | Trichophyton rubrum cDNA library DW706655 | 5e−36 | 3.1 | 6.2 | 11.2 | 3.6 | 3.5 | 99 |
| TR0006 (DW005353) | 250 | A. fumigatus Copper resistance-associated P-type ATPase EAL92309 | 4e−16 | 1.6 | 3.8 | 9.1 | 4.8 | 1.8 | 20 |
| TR0007 (DW005354) | 271 | A. fumigatus Conserved hypothetical protein EAL92616 | 6e−14 | 3.7 | 5.3 | 12.9 | 1.2 | 2.9 | 99 |
| TR0008 (DW005355) | 201 | Chaetomium globosum Hypothetical protein XP_001226392 | 2e−09 | 3.1 | 1.0 | 6.3 | 4.3 | 1.4 | 99 |
| T. rubrum cDNA library DW406176 | 4e−80 | ||||||||
| TR0009 (DW005356) | 326 | Magnaporthe grisea Hypothetical protein XP_367322 | 1e−06 | 3.5 | 3.3 | 4.8 | 4.8 | 4.2 | 99 |
| TR0011 (DW005358) | 337 | A. fumigatus ABC transporter EAL88981 | 1e−20 | 1.8 | 4.0 | 7.5 | 3.5 | 2.3 | 20 |
| TR0012 (DW005359) | 249 | A. fumigatus DNA mismatch repair protein XP_751995 | 1e−33 | 7.2 | 0.8 | 6.8 | 9.3 | 3.1 | 10 |
| TR0013 (DW005360) | 355 | Emericella nidulans NIMA interactive protein AAP23304 | 1e−20 | 1.9 | 15.3 | 16.0 | 3.4 | 2.4 | 10 |
| TR0015 (DW005362) | 486 | Coccidioides immitis Hypothetical protein XP_001247286 | 2e−05 | 2.8 | 11.3 | 5.8 | 6.7 | 3.1 | 99 |
| TR0017 (DW005364) | 190 | Aspergillus terreus DNA polymerase gamma XP_001211183 | 3e−22 | 5.5 | 11.3 | 9.5 | 6.2 | 5.0 | 10 |
| TR0018 (DW005365) | 395 | A. fumigatus Leucine Rich Repeat domain EAL88387 | 3e−13 | 2.9 | 14.0 | 21.0 | 6.0 | 1.6 | 98 |
| TR0020 (DW005367) | 303 | A. fumigatus Hypothetical protein XP_754782 | 2e−05 | 5.2 | 7.3 | 11.9 | 5.0 | 1.9 | 99 |
| TR0023 (DW005370) | 135 | A. fumigatus Hypothetical protein XP_663650 | 3e−05 | 1.6 | 7.0 | 14.0 | 2.9 | 1.7 | 99 |
| TR0024 (DW005371) | 100 | A. nidulans Hypothetical protein XP_662910 | 2e−12 | 4.2 | 4.1 | 9.8 | 7.1 | 5.2 | 99 |
| TR0025 (DW005372) | 228 | C. immitis Glyoxalase/bleomycin resistance XP_001248691 | 7e−23 | 2.2 | 6.3 | 8.8 | 4.0 | 2.7 | 01 |
| TR0027 (DW005374) | 454 | Gibberella zeae Hypothetical protein XP_380291 | 4e−06 | 5.1 | 12.0 | 7.4 | 5.4 | 1.6 | 99 |
| TR0028 (DW005375) | 540 | C. immitis DopAp XP_001247286 | 1e−13 | 5.0 | 12.8 | 5.2 | 11.5 | 6.3 | 40 |
| TR0029 (DW005376) | 241 | Aspergillus oryzae short chain dehydrogenase BAE58435 | 7e−08 | 6.0 | 2.0 | 3.4 | 4.3 | 5.2 | 02 |
| TR0030 (DW005377) | 610 | C. immitis Hypothetical protein XP_001246822 | 5e−40 | 6.6 | 10.1 | 12.9 | 5.0 | 3.2 | 99 |
| T. rubrum cDNA clone AJ883510 | 4e−98 | ||||||||
| TR0031 (DW005378) | 340 | T. rubrum cDNA library DW682179 | 1e−139 | 4.0 | 9.9 | 7.2 | 4.1 | 7.0 | 99 |
| TR0032 (DW005379) | 211 | A. nidulans Hypothetical protein XP_681434 | 3e−08 | 6.1 | 11.0 | 9.7 | 5.0 | 5.3 | 99 |
| T. rubrum cDNA clone AJ883266 | 4e−76 | ||||||||
| TR0033 (DW005380) | 278 | T. rubrum cDNA library DW682095 | 2e−17 | 4.0 | 8.9 | 7.0 | 5.1 | 4.7 | 99 |
| TR0034 (DW005381) | 426 | C. immitis Monooxygenase XP_001243320 | 2e−28 | 4.0 | 6.7 | 11.2 | 5.0 | 3.0 | 02 |
| TR0035 (DW005382) | 233 | C.immitis ATP-dependent RNA helicase XP_001247075 | 4e−15 | 5.2 | 18 | 11 | 6.0 | 2.4 | 11 |
| TR0036 (DW005383) | 299 | A. fumigatus Carboxylic ester hydrolase EAL88020 | 2e−25 | 4.7 | 3.6 | 8.3 | 6.7 | 3.5 | 32 |
| TR0037 (DW005384) | 115 | A. nidulans Hypothetical protein XP_662910 | 4e−11 | 1.7 | 2.0 | 4.0 | 3.1 | 6.8 | 99 |
| TR0038 (DW005385) | 602 | C. globosum nonribosomal peptide synthase XP_001222884 | 7e−07 | 2.3 | 2.4 | 11.0 | 4.0 | 2.0 | 01 |
| TR0039 (AF291822) | 371 | T. rubrum Multidrug resistance protein MDR AAG01549 | 2e−21 | 4.6 | 13.1 | 10.0 | 6.4 | 5.1 | 20 |
| TR0010 (DW005357) | 154 | No significant matches | 6.5 | 12.8 | 8.5 | 5.3 | 3.6 | 99 | |
| TR0014 (DW005361) | 198 | No significant matches | 4.0 | 20.0 | 3.2 | 7.7 | 4.3 | 99 | |
| TR0016 (DW005363) | 198 | No significant matches | 2.9 | 6.5 | 9.6 | 12.0 | 5.7 | 99 | |
| TR0019 (DW005366) | 235 | No significant matches | 2.6 | 10.0 | 4.4 | 4.1 | 3.4 | 99 | |
| TR0021 (DW005368) | 222 | No significant matches | 7.1 | 9.7 | 10.0 | 7.3 | 7.9 | 99 | |
| TR0022 (DW005369) | 303 | No significant matches | 3.4 | 1.7 | 5.5 | 3.8 | 4.1 | 99 | |
| TR0026 (DW005373) | 158 | No significant matches | 5.0 | 10.9 | 11.2 | 5.1 | 4.7 | 99 | |
| Change in fold expression | |||||||||
| Clones(GenBank Accession No.) | Size (bp) | Putative ID | E-value | ACR | FLC | GRS | TRB | UDA | MIPs functional category |
| TR0001 (DW005348) | 578 | Aspergillus nidulans Hypothetical protein XP_663997 | 8e−05 | 3.1 | 6.0 | 3.5 | 10.0 | 4.3 | 99 |
| TR0002 (DW005349) | 415 | Glomerella cingulata Pol protein AF264028 | 1e−42 | 3.1 | 10.9 | 6.0 | 2.9 | 1.1 | 38 |
| TR0003 (DW005350) | 156 | Aspergillus fumigatus DEAD helicases superfamily EAL84582 | 8e−20 | 7.9 | 13.4 | 6.0 | 7.9 | 4.8 | 11 |
| TR0004 (DW005351) | 229 | A. fumigatus Kynurenine aminotransferase XP_751710 | 3e−28 | 0.6 | 6.3 | 1.2 | 4.0 | 1.9 | 01 |
| TR0005 (DW005352) | 197 | Trichophyton rubrum cDNA library DW706655 | 5e−36 | 3.1 | 6.2 | 11.2 | 3.6 | 3.5 | 99 |
| TR0006 (DW005353) | 250 | A. fumigatus Copper resistance-associated P-type ATPase EAL92309 | 4e−16 | 1.6 | 3.8 | 9.1 | 4.8 | 1.8 | 20 |
| TR0007 (DW005354) | 271 | A. fumigatus Conserved hypothetical protein EAL92616 | 6e−14 | 3.7 | 5.3 | 12.9 | 1.2 | 2.9 | 99 |
| TR0008 (DW005355) | 201 | Chaetomium globosum Hypothetical protein XP_001226392 | 2e−09 | 3.1 | 1.0 | 6.3 | 4.3 | 1.4 | 99 |
| T. rubrum cDNA library DW406176 | 4e−80 | ||||||||
| TR0009 (DW005356) | 326 | Magnaporthe grisea Hypothetical protein XP_367322 | 1e−06 | 3.5 | 3.3 | 4.8 | 4.8 | 4.2 | 99 |
| TR0011 (DW005358) | 337 | A. fumigatus ABC transporter EAL88981 | 1e−20 | 1.8 | 4.0 | 7.5 | 3.5 | 2.3 | 20 |
| TR0012 (DW005359) | 249 | A. fumigatus DNA mismatch repair protein XP_751995 | 1e−33 | 7.2 | 0.8 | 6.8 | 9.3 | 3.1 | 10 |
| TR0013 (DW005360) | 355 | Emericella nidulans NIMA interactive protein AAP23304 | 1e−20 | 1.9 | 15.3 | 16.0 | 3.4 | 2.4 | 10 |
| TR0015 (DW005362) | 486 | Coccidioides immitis Hypothetical protein XP_001247286 | 2e−05 | 2.8 | 11.3 | 5.8 | 6.7 | 3.1 | 99 |
| TR0017 (DW005364) | 190 | Aspergillus terreus DNA polymerase gamma XP_001211183 | 3e−22 | 5.5 | 11.3 | 9.5 | 6.2 | 5.0 | 10 |
| TR0018 (DW005365) | 395 | A. fumigatus Leucine Rich Repeat domain EAL88387 | 3e−13 | 2.9 | 14.0 | 21.0 | 6.0 | 1.6 | 98 |
| TR0020 (DW005367) | 303 | A. fumigatus Hypothetical protein XP_754782 | 2e−05 | 5.2 | 7.3 | 11.9 | 5.0 | 1.9 | 99 |
| TR0023 (DW005370) | 135 | A. fumigatus Hypothetical protein XP_663650 | 3e−05 | 1.6 | 7.0 | 14.0 | 2.9 | 1.7 | 99 |
| TR0024 (DW005371) | 100 | A. nidulans Hypothetical protein XP_662910 | 2e−12 | 4.2 | 4.1 | 9.8 | 7.1 | 5.2 | 99 |
| TR0025 (DW005372) | 228 | C. immitis Glyoxalase/bleomycin resistance XP_001248691 | 7e−23 | 2.2 | 6.3 | 8.8 | 4.0 | 2.7 | 01 |
| TR0027 (DW005374) | 454 | Gibberella zeae Hypothetical protein XP_380291 | 4e−06 | 5.1 | 12.0 | 7.4 | 5.4 | 1.6 | 99 |
| TR0028 (DW005375) | 540 | C. immitis DopAp XP_001247286 | 1e−13 | 5.0 | 12.8 | 5.2 | 11.5 | 6.3 | 40 |
| TR0029 (DW005376) | 241 | Aspergillus oryzae short chain dehydrogenase BAE58435 | 7e−08 | 6.0 | 2.0 | 3.4 | 4.3 | 5.2 | 02 |
| TR0030 (DW005377) | 610 | C. immitis Hypothetical protein XP_001246822 | 5e−40 | 6.6 | 10.1 | 12.9 | 5.0 | 3.2 | 99 |
| T. rubrum cDNA clone AJ883510 | 4e−98 | ||||||||
| TR0031 (DW005378) | 340 | T. rubrum cDNA library DW682179 | 1e−139 | 4.0 | 9.9 | 7.2 | 4.1 | 7.0 | 99 |
| TR0032 (DW005379) | 211 | A. nidulans Hypothetical protein XP_681434 | 3e−08 | 6.1 | 11.0 | 9.7 | 5.0 | 5.3 | 99 |
| T. rubrum cDNA clone AJ883266 | 4e−76 | ||||||||
| TR0033 (DW005380) | 278 | T. rubrum cDNA library DW682095 | 2e−17 | 4.0 | 8.9 | 7.0 | 5.1 | 4.7 | 99 |
| TR0034 (DW005381) | 426 | C. immitis Monooxygenase XP_001243320 | 2e−28 | 4.0 | 6.7 | 11.2 | 5.0 | 3.0 | 02 |
| TR0035 (DW005382) | 233 | C.immitis ATP-dependent RNA helicase XP_001247075 | 4e−15 | 5.2 | 18 | 11 | 6.0 | 2.4 | 11 |
| TR0036 (DW005383) | 299 | A. fumigatus Carboxylic ester hydrolase EAL88020 | 2e−25 | 4.7 | 3.6 | 8.3 | 6.7 | 3.5 | 32 |
| TR0037 (DW005384) | 115 | A. nidulans Hypothetical protein XP_662910 | 4e−11 | 1.7 | 2.0 | 4.0 | 3.1 | 6.8 | 99 |
| TR0038 (DW005385) | 602 | C. globosum nonribosomal peptide synthase XP_001222884 | 7e−07 | 2.3 | 2.4 | 11.0 | 4.0 | 2.0 | 01 |
| TR0039 (AF291822) | 371 | T. rubrum Multidrug resistance protein MDR AAG01549 | 2e−21 | 4.6 | 13.1 | 10.0 | 6.4 | 5.1 | 20 |
| TR0010 (DW005357) | 154 | No significant matches | 6.5 | 12.8 | 8.5 | 5.3 | 3.6 | 99 | |
| TR0014 (DW005361) | 198 | No significant matches | 4.0 | 20.0 | 3.2 | 7.7 | 4.3 | 99 | |
| TR0016 (DW005363) | 198 | No significant matches | 2.9 | 6.5 | 9.6 | 12.0 | 5.7 | 99 | |
| TR0019 (DW005366) | 235 | No significant matches | 2.6 | 10.0 | 4.4 | 4.1 | 3.4 | 99 | |
| TR0021 (DW005368) | 222 | No significant matches | 7.1 | 9.7 | 10.0 | 7.3 | 7.9 | 99 | |
| TR0022 (DW005369) | 303 | No significant matches | 3.4 | 1.7 | 5.5 | 3.8 | 4.1 | 99 | |
| TR0026 (DW005373) | 158 | No significant matches | 5.0 | 10.9 | 11.2 | 5.1 | 4.7 | 99 | |
Putative identification is based on blast searches of the nonredundant GenBank database. Only scores with an E-value of <10e−3 were considered significant.
Insert size sequenced with quality ≥20 attributed by phred software.
The ratio of the cDNA clones signal intensities from each drug (acriflavin, ACR; fluconazole, FLC; griseofulfin, GRS; terbinafine, TRB; or undecanoic acid, UDA) to control samples generated from macro-array dot-blot assays. In bold are the intensities of hybridization fourfold or above. MIPS: 1, metabolism; 2, energy; 10, cell cycle/DNA processing; 11, transcription; 20, transport facilitation; 32, cell rescue/defense/virulence; 38, transposable element; 40, cell fate; 98 and 99, uncharacterized.
GenBank est database.
Expression and homologous sequences of putative genes from Trichophyton rubrum SSH library
| Change in fold expression | |||||||||
| Clones(GenBank Accession No.) | Size (bp) | Putative ID | E-value | ACR | FLC | GRS | TRB | UDA | MIPs functional category |
| TR0001 (DW005348) | 578 | Aspergillus nidulans Hypothetical protein XP_663997 | 8e−05 | 3.1 | 6.0 | 3.5 | 10.0 | 4.3 | 99 |
| TR0002 (DW005349) | 415 | Glomerella cingulata Pol protein AF264028 | 1e−42 | 3.1 | 10.9 | 6.0 | 2.9 | 1.1 | 38 |
| TR0003 (DW005350) | 156 | Aspergillus fumigatus DEAD helicases superfamily EAL84582 | 8e−20 | 7.9 | 13.4 | 6.0 | 7.9 | 4.8 | 11 |
| TR0004 (DW005351) | 229 | A. fumigatus Kynurenine aminotransferase XP_751710 | 3e−28 | 0.6 | 6.3 | 1.2 | 4.0 | 1.9 | 01 |
| TR0005 (DW005352) | 197 | Trichophyton rubrum cDNA library DW706655 | 5e−36 | 3.1 | 6.2 | 11.2 | 3.6 | 3.5 | 99 |
| TR0006 (DW005353) | 250 | A. fumigatus Copper resistance-associated P-type ATPase EAL92309 | 4e−16 | 1.6 | 3.8 | 9.1 | 4.8 | 1.8 | 20 |
| TR0007 (DW005354) | 271 | A. fumigatus Conserved hypothetical protein EAL92616 | 6e−14 | 3.7 | 5.3 | 12.9 | 1.2 | 2.9 | 99 |
| TR0008 (DW005355) | 201 | Chaetomium globosum Hypothetical protein XP_001226392 | 2e−09 | 3.1 | 1.0 | 6.3 | 4.3 | 1.4 | 99 |
| T. rubrum cDNA library DW406176 | 4e−80 | ||||||||
| TR0009 (DW005356) | 326 | Magnaporthe grisea Hypothetical protein XP_367322 | 1e−06 | 3.5 | 3.3 | 4.8 | 4.8 | 4.2 | 99 |
| TR0011 (DW005358) | 337 | A. fumigatus ABC transporter EAL88981 | 1e−20 | 1.8 | 4.0 | 7.5 | 3.5 | 2.3 | 20 |
| TR0012 (DW005359) | 249 | A. fumigatus DNA mismatch repair protein XP_751995 | 1e−33 | 7.2 | 0.8 | 6.8 | 9.3 | 3.1 | 10 |
| TR0013 (DW005360) | 355 | Emericella nidulans NIMA interactive protein AAP23304 | 1e−20 | 1.9 | 15.3 | 16.0 | 3.4 | 2.4 | 10 |
| TR0015 (DW005362) | 486 | Coccidioides immitis Hypothetical protein XP_001247286 | 2e−05 | 2.8 | 11.3 | 5.8 | 6.7 | 3.1 | 99 |
| TR0017 (DW005364) | 190 | Aspergillus terreus DNA polymerase gamma XP_001211183 | 3e−22 | 5.5 | 11.3 | 9.5 | 6.2 | 5.0 | 10 |
| TR0018 (DW005365) | 395 | A. fumigatus Leucine Rich Repeat domain EAL88387 | 3e−13 | 2.9 | 14.0 | 21.0 | 6.0 | 1.6 | 98 |
| TR0020 (DW005367) | 303 | A. fumigatus Hypothetical protein XP_754782 | 2e−05 | 5.2 | 7.3 | 11.9 | 5.0 | 1.9 | 99 |
| TR0023 (DW005370) | 135 | A. fumigatus Hypothetical protein XP_663650 | 3e−05 | 1.6 | 7.0 | 14.0 | 2.9 | 1.7 | 99 |
| TR0024 (DW005371) | 100 | A. nidulans Hypothetical protein XP_662910 | 2e−12 | 4.2 | 4.1 | 9.8 | 7.1 | 5.2 | 99 |
| TR0025 (DW005372) | 228 | C. immitis Glyoxalase/bleomycin resistance XP_001248691 | 7e−23 | 2.2 | 6.3 | 8.8 | 4.0 | 2.7 | 01 |
| TR0027 (DW005374) | 454 | Gibberella zeae Hypothetical protein XP_380291 | 4e−06 | 5.1 | 12.0 | 7.4 | 5.4 | 1.6 | 99 |
| TR0028 (DW005375) | 540 | C. immitis DopAp XP_001247286 | 1e−13 | 5.0 | 12.8 | 5.2 | 11.5 | 6.3 | 40 |
| TR0029 (DW005376) | 241 | Aspergillus oryzae short chain dehydrogenase BAE58435 | 7e−08 | 6.0 | 2.0 | 3.4 | 4.3 | 5.2 | 02 |
| TR0030 (DW005377) | 610 | C. immitis Hypothetical protein XP_001246822 | 5e−40 | 6.6 | 10.1 | 12.9 | 5.0 | 3.2 | 99 |
| T. rubrum cDNA clone AJ883510 | 4e−98 | ||||||||
| TR0031 (DW005378) | 340 | T. rubrum cDNA library DW682179 | 1e−139 | 4.0 | 9.9 | 7.2 | 4.1 | 7.0 | 99 |
| TR0032 (DW005379) | 211 | A. nidulans Hypothetical protein XP_681434 | 3e−08 | 6.1 | 11.0 | 9.7 | 5.0 | 5.3 | 99 |
| T. rubrum cDNA clone AJ883266 | 4e−76 | ||||||||
| TR0033 (DW005380) | 278 | T. rubrum cDNA library DW682095 | 2e−17 | 4.0 | 8.9 | 7.0 | 5.1 | 4.7 | 99 |
| TR0034 (DW005381) | 426 | C. immitis Monooxygenase XP_001243320 | 2e−28 | 4.0 | 6.7 | 11.2 | 5.0 | 3.0 | 02 |
| TR0035 (DW005382) | 233 | C.immitis ATP-dependent RNA helicase XP_001247075 | 4e−15 | 5.2 | 18 | 11 | 6.0 | 2.4 | 11 |
| TR0036 (DW005383) | 299 | A. fumigatus Carboxylic ester hydrolase EAL88020 | 2e−25 | 4.7 | 3.6 | 8.3 | 6.7 | 3.5 | 32 |
| TR0037 (DW005384) | 115 | A. nidulans Hypothetical protein XP_662910 | 4e−11 | 1.7 | 2.0 | 4.0 | 3.1 | 6.8 | 99 |
| TR0038 (DW005385) | 602 | C. globosum nonribosomal peptide synthase XP_001222884 | 7e−07 | 2.3 | 2.4 | 11.0 | 4.0 | 2.0 | 01 |
| TR0039 (AF291822) | 371 | T. rubrum Multidrug resistance protein MDR AAG01549 | 2e−21 | 4.6 | 13.1 | 10.0 | 6.4 | 5.1 | 20 |
| TR0010 (DW005357) | 154 | No significant matches | 6.5 | 12.8 | 8.5 | 5.3 | 3.6 | 99 | |
| TR0014 (DW005361) | 198 | No significant matches | 4.0 | 20.0 | 3.2 | 7.7 | 4.3 | 99 | |
| TR0016 (DW005363) | 198 | No significant matches | 2.9 | 6.5 | 9.6 | 12.0 | 5.7 | 99 | |
| TR0019 (DW005366) | 235 | No significant matches | 2.6 | 10.0 | 4.4 | 4.1 | 3.4 | 99 | |
| TR0021 (DW005368) | 222 | No significant matches | 7.1 | 9.7 | 10.0 | 7.3 | 7.9 | 99 | |
| TR0022 (DW005369) | 303 | No significant matches | 3.4 | 1.7 | 5.5 | 3.8 | 4.1 | 99 | |
| TR0026 (DW005373) | 158 | No significant matches | 5.0 | 10.9 | 11.2 | 5.1 | 4.7 | 99 | |
| Change in fold expression | |||||||||
| Clones(GenBank Accession No.) | Size (bp) | Putative ID | E-value | ACR | FLC | GRS | TRB | UDA | MIPs functional category |
| TR0001 (DW005348) | 578 | Aspergillus nidulans Hypothetical protein XP_663997 | 8e−05 | 3.1 | 6.0 | 3.5 | 10.0 | 4.3 | 99 |
| TR0002 (DW005349) | 415 | Glomerella cingulata Pol protein AF264028 | 1e−42 | 3.1 | 10.9 | 6.0 | 2.9 | 1.1 | 38 |
| TR0003 (DW005350) | 156 | Aspergillus fumigatus DEAD helicases superfamily EAL84582 | 8e−20 | 7.9 | 13.4 | 6.0 | 7.9 | 4.8 | 11 |
| TR0004 (DW005351) | 229 | A. fumigatus Kynurenine aminotransferase XP_751710 | 3e−28 | 0.6 | 6.3 | 1.2 | 4.0 | 1.9 | 01 |
| TR0005 (DW005352) | 197 | Trichophyton rubrum cDNA library DW706655 | 5e−36 | 3.1 | 6.2 | 11.2 | 3.6 | 3.5 | 99 |
| TR0006 (DW005353) | 250 | A. fumigatus Copper resistance-associated P-type ATPase EAL92309 | 4e−16 | 1.6 | 3.8 | 9.1 | 4.8 | 1.8 | 20 |
| TR0007 (DW005354) | 271 | A. fumigatus Conserved hypothetical protein EAL92616 | 6e−14 | 3.7 | 5.3 | 12.9 | 1.2 | 2.9 | 99 |
| TR0008 (DW005355) | 201 | Chaetomium globosum Hypothetical protein XP_001226392 | 2e−09 | 3.1 | 1.0 | 6.3 | 4.3 | 1.4 | 99 |
| T. rubrum cDNA library DW406176 | 4e−80 | ||||||||
| TR0009 (DW005356) | 326 | Magnaporthe grisea Hypothetical protein XP_367322 | 1e−06 | 3.5 | 3.3 | 4.8 | 4.8 | 4.2 | 99 |
| TR0011 (DW005358) | 337 | A. fumigatus ABC transporter EAL88981 | 1e−20 | 1.8 | 4.0 | 7.5 | 3.5 | 2.3 | 20 |
| TR0012 (DW005359) | 249 | A. fumigatus DNA mismatch repair protein XP_751995 | 1e−33 | 7.2 | 0.8 | 6.8 | 9.3 | 3.1 | 10 |
| TR0013 (DW005360) | 355 | Emericella nidulans NIMA interactive protein AAP23304 | 1e−20 | 1.9 | 15.3 | 16.0 | 3.4 | 2.4 | 10 |
| TR0015 (DW005362) | 486 | Coccidioides immitis Hypothetical protein XP_001247286 | 2e−05 | 2.8 | 11.3 | 5.8 | 6.7 | 3.1 | 99 |
| TR0017 (DW005364) | 190 | Aspergillus terreus DNA polymerase gamma XP_001211183 | 3e−22 | 5.5 | 11.3 | 9.5 | 6.2 | 5.0 | 10 |
| TR0018 (DW005365) | 395 | A. fumigatus Leucine Rich Repeat domain EAL88387 | 3e−13 | 2.9 | 14.0 | 21.0 | 6.0 | 1.6 | 98 |
| TR0020 (DW005367) | 303 | A. fumigatus Hypothetical protein XP_754782 | 2e−05 | 5.2 | 7.3 | 11.9 | 5.0 | 1.9 | 99 |
| TR0023 (DW005370) | 135 | A. fumigatus Hypothetical protein XP_663650 | 3e−05 | 1.6 | 7.0 | 14.0 | 2.9 | 1.7 | 99 |
| TR0024 (DW005371) | 100 | A. nidulans Hypothetical protein XP_662910 | 2e−12 | 4.2 | 4.1 | 9.8 | 7.1 | 5.2 | 99 |
| TR0025 (DW005372) | 228 | C. immitis Glyoxalase/bleomycin resistance XP_001248691 | 7e−23 | 2.2 | 6.3 | 8.8 | 4.0 | 2.7 | 01 |
| TR0027 (DW005374) | 454 | Gibberella zeae Hypothetical protein XP_380291 | 4e−06 | 5.1 | 12.0 | 7.4 | 5.4 | 1.6 | 99 |
| TR0028 (DW005375) | 540 | C. immitis DopAp XP_001247286 | 1e−13 | 5.0 | 12.8 | 5.2 | 11.5 | 6.3 | 40 |
| TR0029 (DW005376) | 241 | Aspergillus oryzae short chain dehydrogenase BAE58435 | 7e−08 | 6.0 | 2.0 | 3.4 | 4.3 | 5.2 | 02 |
| TR0030 (DW005377) | 610 | C. immitis Hypothetical protein XP_001246822 | 5e−40 | 6.6 | 10.1 | 12.9 | 5.0 | 3.2 | 99 |
| T. rubrum cDNA clone AJ883510 | 4e−98 | ||||||||
| TR0031 (DW005378) | 340 | T. rubrum cDNA library DW682179 | 1e−139 | 4.0 | 9.9 | 7.2 | 4.1 | 7.0 | 99 |
| TR0032 (DW005379) | 211 | A. nidulans Hypothetical protein XP_681434 | 3e−08 | 6.1 | 11.0 | 9.7 | 5.0 | 5.3 | 99 |
| T. rubrum cDNA clone AJ883266 | 4e−76 | ||||||||
| TR0033 (DW005380) | 278 | T. rubrum cDNA library DW682095 | 2e−17 | 4.0 | 8.9 | 7.0 | 5.1 | 4.7 | 99 |
| TR0034 (DW005381) | 426 | C. immitis Monooxygenase XP_001243320 | 2e−28 | 4.0 | 6.7 | 11.2 | 5.0 | 3.0 | 02 |
| TR0035 (DW005382) | 233 | C.immitis ATP-dependent RNA helicase XP_001247075 | 4e−15 | 5.2 | 18 | 11 | 6.0 | 2.4 | 11 |
| TR0036 (DW005383) | 299 | A. fumigatus Carboxylic ester hydrolase EAL88020 | 2e−25 | 4.7 | 3.6 | 8.3 | 6.7 | 3.5 | 32 |
| TR0037 (DW005384) | 115 | A. nidulans Hypothetical protein XP_662910 | 4e−11 | 1.7 | 2.0 | 4.0 | 3.1 | 6.8 | 99 |
| TR0038 (DW005385) | 602 | C. globosum nonribosomal peptide synthase XP_001222884 | 7e−07 | 2.3 | 2.4 | 11.0 | 4.0 | 2.0 | 01 |
| TR0039 (AF291822) | 371 | T. rubrum Multidrug resistance protein MDR AAG01549 | 2e−21 | 4.6 | 13.1 | 10.0 | 6.4 | 5.1 | 20 |
| TR0010 (DW005357) | 154 | No significant matches | 6.5 | 12.8 | 8.5 | 5.3 | 3.6 | 99 | |
| TR0014 (DW005361) | 198 | No significant matches | 4.0 | 20.0 | 3.2 | 7.7 | 4.3 | 99 | |
| TR0016 (DW005363) | 198 | No significant matches | 2.9 | 6.5 | 9.6 | 12.0 | 5.7 | 99 | |
| TR0019 (DW005366) | 235 | No significant matches | 2.6 | 10.0 | 4.4 | 4.1 | 3.4 | 99 | |
| TR0021 (DW005368) | 222 | No significant matches | 7.1 | 9.7 | 10.0 | 7.3 | 7.9 | 99 | |
| TR0022 (DW005369) | 303 | No significant matches | 3.4 | 1.7 | 5.5 | 3.8 | 4.1 | 99 | |
| TR0026 (DW005373) | 158 | No significant matches | 5.0 | 10.9 | 11.2 | 5.1 | 4.7 | 99 | |
Putative identification is based on blast searches of the nonredundant GenBank database. Only scores with an E-value of <10e−3 were considered significant.
Insert size sequenced with quality ≥20 attributed by phred software.
The ratio of the cDNA clones signal intensities from each drug (acriflavin, ACR; fluconazole, FLC; griseofulfin, GRS; terbinafine, TRB; or undecanoic acid, UDA) to control samples generated from macro-array dot-blot assays. In bold are the intensities of hybridization fourfold or above. MIPS: 1, metabolism; 2, energy; 10, cell cycle/DNA processing; 11, transcription; 20, transport facilitation; 32, cell rescue/defense/virulence; 38, transposable element; 40, cell fate; 98 and 99, uncharacterized.
GenBank est database.
Dot-blot and reverse Northern experiments
cDNA inserts in individual E. coli colonies were analyzed by PCR. Two microliters of overnight culture was added to a master mix containing 1 mM nested primers 1 e 2R (Clontech PCR-Select cDNA Subtraction Kit), 1.5 mM MgCl2, 200 µM each deoxynucleotide, 1 U of Taq DNA polymerase and PCR buffer supplied by the manufacturer (Invitrogen). PCR was performed using the Applied Biosystem GeneAmp 9700 PCR system according to the following parameters: 94°C for 5 min, and 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 1 min. All PCR products were checked on 0.8% agarose gel stained with ethidium bromide. Identical quantities of the PCR products were denatured with 0.6 N NaOH before being spotted onto Hybond-N+Nylon membranes using Bio-Dot apparatus (Bio-Rad). Six complex cDNA probes were made using 5 µg RNA extracted from H6 not exposed to drugs and 5 µg RNA extracted from H6 exposed to each different inhibitor agent (acriflavin, fluconazole, griseofulvin, terbinafine and undecanoic acid). The cDNA was obtained using oligo (dT) primer and MML-V reverse transcriptase (Promega) according to the manufacturer's instructions. One microliter of cDNA was employed for probe synthesis by Gene Image random primer labeling module and Gene Image CDP-Star detection module kits (Amersham Biosciences). The membranes were probed at 60°C for 18 h. After washes, following the manufacturer's instruction, the membranes were exposed to autoradiography for 2 h at room temperature. The membranes were firstly probed with cDNA population from H6 not exposed to drugs (control) and then reprobed (after stripping the first probe) with cDNA population from H6 exposed to each drug.
For densiometric analysis, hybridization signals were quantified with the image j software (Research Services Branch, National Institute of Mental Health, Bethesda, MD, wayne@codon.nih.gov). A fourfold increase in the densiometric values of each spot obtained from T. rubrum cDNA, submitted to each drug treatment and compared with the signal in the untreated cultures, was arbitrarily considered significant.
Northern blot analysis
For validation of differential gene expression by Northern blot, T. rubrum was cultivated in the presence of the antifungal agents. Total RNA from H6 exposed to different drugs (15 min at 90% of their MICs, 30 or 60 min at 75% of their MICs) were extracted, and c. 15 µg was electrophoresed on 1.5% agarose gel containing formaldehyde, transferred onto a Hybond-N+membrane and hybridized with subtracted clones amplified by PCR radioactively labeled with [α-32P]-dCTP, as previously described (Sambrook et al., 1989).
Results
Confirmation of differential expression by macro-array dot-blot
A subtractive library containing 204 clones enriched for T. rubrum genes up-regulated under cytotoxic agents exposure was constructed. To identify which bacterial colonies possessed cloning vectors with inserts, a nested PCR was performed, using nested primer 1 and 2R included with the SSH kit. Insert screening analysis reveals that the amplified cDNA inserts ranged in size from 0.25 to 1.3 kb, showing that those clones amplified had cDNA inserts carrying adaptor 1 and adaptor 2R on both ends, which suggests that contaminating background due to nonspecific amplification of the tester was low. Of the 204 colonies analyzed, 143 had inserts and equal amounts of amplified cDNA of these positive clones were arrayed on two nylon membranes, in duplicate, for a total of four membranes. Each membrane was hybridized with six different probes. Firstly, the total cDNA population probe, obtained from H6 strain cultured into drug-free medium, was used as a negative control. Then, the membranes were successively hybridized with cDNA population probes obtained from H6 strain exposed to each cytotoxic compound. Figure 1 shows a representative subset of clones submitted to this macro-array dot-blot screening. Clones that hybridized strongly to the cDNA probes obtained from H6 strain challenged with cytotoxic drugs, compared with the control, were considered candidates for differential expression.
Macro-array dot-blot. Membranes blotted with amplified cDNA sequences from subtracted library, probed with total cDNA population from Trichophyton rubrum cultured into drug-free medium (a) and reprobed with total cDNA population from T. rubrum exposed to fluconazole (b). Differentially expressed transcripts obtained from T. rubrum challenged with cytotoxic drugs represent the clones with stronger signals (b) compared to control (a).
Sequence and Northern analyses
Among the 143 subtractive cDNA clones screened, 132 were sequenced because they correspond to genes differentially expressed after T. rubrum exposure to at least one of the cytotoxic drugs. Thirty-nine nonredundant unique ESTs were generated and compared with NCBI GenBank database using the blast algorithm (Table 1). Sequence analysis revealed that among the 39 putative upregulated T. rubrum ESTs, 17 matched known genes and 12 matched hypothetical proteins. The remainder could be unique to T. rubrum and other dermatophytes. Of the 32 genes that had significant hits in the GenBank, 100% had similarity to sequences from fungi.
All the 39 ESTs were induced by more than one drug (Table 1). These results suggest that most of the genes revealed in this study could be involved in nonspecific responses to cellular stress.
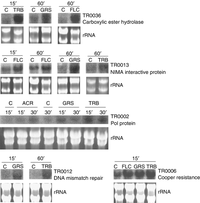
The upregulation of the T. rubrum genes coding for Pol protein (TR0002), Copper resistance-associated P-type ATPase (TR0006), DNA mismatch repair protein (TR0012), NIMA (never in mitosis A) interactive protein (TR0013) and Carboxylic ester hydrolase (TR0036) in response to some cytotoxic drugs was confirmed by Northern blot analyses (Fig. 2).
Northern blot analyses of five genes using total RNA from strain H6 cultured in liquid Sabouraud medium for 72 h and treated with the following toxicants for 15, 30 or 60 min: TRB (terbinafine), GRS (griseofulvin), FLC (fluconazole) or ACR (acriflavin). C: control. Ethidium bromide-stained rRNA bands are shown for comparison of the quantities of loaded RNA. The identification and functional description of the clones is shown in Table 1.
Discussion
Drugs provoke cellular stress, eliciting adaptive responses that allow the cells to overcome the toxic effects of these chemicals (Hayes & Wolf, 1990). This ability to resist chemical challenge can arise immediately following drug administration. The purpose of this study was to identify T. rubrum genes upregulated during exposure to cytotoxic drugs with the hypothesis that genes expressed upon exposure to drugs can help to understand how the adaptation to physiological stress occurs and how these drugs work in this fungus. The SSH technique revealed 39 T. rubrum genes upregulated during the exposure to acriflavin, fluconazole, griseofulvin, terbinafine and/or undecanoic acid, which were screened by macro-array dot-blot. Among the 39 genes, 32 were novel T. rubrum genes, representing an increase in the number of T. rubrum genes that have been reported. One gene (TruMDR2) that had been previously reported (TR0039, AF291822) to code for a putative Multidrug resistance protein was upregulated after exposure of T. rubrum to all of the drugs assayed. In fact, this energy-dependent drug efflux mechanism is implicated in resistance to a wide range of unrelated cytotoxic compounds. Northern analysis of this gene revealed an increased level of transcription when mycelia were exposed to acriflavin, griseofulvin or fluconazole (Fachin et al., 2006), validating the results obtained by macro-array dot-blot. Also, disruption of the TruMDR2 gene rendered the mutant more sensitive to terbinafine than the wild type, suggesting that this transporter plays a role in modulating drug susceptibility in T. rubrum (Fachin et al., 2006). Another cDNA isolated by subtraction experiments (TR0011) also encodes a predicted transporter similar to MDR-like proteins in Aspergillus fumigatus (EAL88981), Aspergillus orizae (BAE59019) and Aspergillus nidulans (EAA66660), which do not have their physiological roles described. In T. rubrum, this gene (TR0011) is upregulated in response to fluconazole or griseofulvin, suggesting that it could be a potential determinant for an MDR phenotype.
Another EST upregulated upon treatment of T. rubrum with griseofulvin, terbinafine and more weakly with fluconazole (TR0006) encodes a transporter similar to a copper resistance-associated P-type ATPase protein. Copper is an essential redox-active cofactor for proteins involved in energy generation, iron uptake and distribution, protection against oxidative stress and many other physiological processes such as virulence (Rees & Thiele, 2004). However, high copper concentrations are toxic and copper resistance is achieved by a variety of mechanisms including the extrusion pumps of P-type ATPase family of transporters (Diatchenko et al., 1996; Weissman et al., 2000). Thus, it seems possible that the TR0006 protein is involved in the maintenance of copper homeostasis, and Northern blot analysis also showed its involvement in the response of T. rubrum towards drug exposure (Fig. 2).
Other ESTs matching genes was found whose products are probably involved in cellular stress and virulence. The predicted products of these genes include a carboxylic ester hydrolase (TR0036) and a Pol protein (TR0002). The gene that encodes the carboxylic ester hydrolase was differentially expressed in all treatments, except under undecanoic acid. The differential expression of this gene was also confirmed by Northern blot when T. rubrum was submitted to fluconazole, griseofulvin or terbinafine (Fig. 2). Although the physiological role of the esterases is not clear, it is known that these enzymes are secreted by T. rubrum (Brasch & Zaldua, 1994). In addition, extra bands in the electrophoretic pattern of T. rubrum intracellular esterase were detected when this fungus was grown in the presence of subinhibitory concentrations of the antimycotics tioconazole or griseofulvin (Fachin et al., 2001). Thus, upregulation of genes encoding esterases may be a nonspecific response to the cellular stress caused by drug challenge. This phenomenon modulates the expression of various virulence mechanisms in bacteria (Held et al., 1995).
The identification of ESTs similar to pol gene of Cgret retrotransposon element from Glomerella cingulata (anamorph: Colletotrichum gloeosporioides) suggests that a transposable element (TE) participates in T. rubrum drug stress response. In fact, transposition in response to environmental stress has been proposed as an adaptive response of the genome. Various transposons in plants, yeasts and Drosophila have been shown to be activated under stress conditions (Eto et al., 2001; Daboussi & Capy, 2003). This EST (TR0002) was shown by macro-array dot-blot to be upregulated in response to fluconazole and griseofulvin antimycotics, but the Northern blot revealed that it is upregulated also in response to acriflavin and terbinafine (Fig. 2). In spite of the fact that retrotransposons had been isolated from other pathogenic fungi, it is the first time that the retrotransposon activity was observed in dermatophytes and the first evidence that drug stress is associated with TE activation.
Also, the predicted products of ESTs TR0012, TR0013 and TR0017 that have similarity to DNA mismatch repair protein, NIMA interactive protein and DNA polymerase gamma, respectively, were classified by MIPs into Cell cycle/DNA-processing category and may be responsive to cellular stresses (Noguchi et al., 2002). The upregulation of the gene encoding the DNA mismatch repair protein in response to griseofulvin and terbinafine, and of the gene encoding NIMA interactive protein in response to fluconazole, griseofulvin and terbinafine was confirmed by Northern blot analyses (Fig. 2).
Taken together, results of this study show that when T. rubrum, grown in Sabouraud medium, is exposed for a short time to a subinhibitory dose of acriflavin, fluconazole, griseofulvin, terbinafine or undecanoic acid, it responds by inducing genes, which suggest novel effects of these drugs. In addition, responses shared across multiple classes of cytotoxic agents were identified, suggesting nonspecific responses to drug challenge, leading to stress adaptation.
Acknowledgements
This work was supported by grants from the Brazilian funding agencies FAPESP, CNPq, CAPES and FAEPA. A. Borghi is thanked for the English manuscript review, and P.R. Sanches, R.A. Ferreira, A.C.C. Souza and M. Mazucato for technical assistance.
References
Author notes
Editor: Derek Jamieson