-
PDF
- Split View
-
Views
-
Cite
Cite
Yutaro Neriya, Kyoko Sugawara, Kensaku Maejima, Masayoshi Hashimoto, Ken Komatsu, Nami Minato, Chihiro Miura, Shigeyuki Kakizawa, Yasuyuki Yamaji, Kenro Oshima, Shigetou Namba, Cloning, expression analysis, and sequence diversity of genes encoding two different immunodominant membrane proteins in poinsettia branch-inducing phytoplasma (PoiBI), FEMS Microbiology Letters, Volume 324, Issue 1, November 2011, Pages 38–47, https://doi.org/10.1111/j.1574-6968.2011.02384.x
Close - Share Icon Share
Abstract
Poinsettia branch-inducing phytoplasma (PoiBI) is a phytopathogenic bacterium that infects poinsettia, and is associated with the free-branching morphotype (characterized by many axillary shoots and flowers) of many commercially grown poinsettias. The major membrane proteins of phytoplasmas are classified into three general types, that is, immunodominant membrane protein (Imp), immunodominant membrane protein A (IdpA), and antigenic membrane protein (Amp). These membrane proteins are often used as targets for the production of antibodies used in phytoplasma detection. Herein, we cloned and sequenced the imp and idpA genes of PoiBI strains from 26 commercial poinsettia cultivars. Although the amino acid sequences of the encoded IdpA proteins were invariant, those of the encoded Imp varied among the PoiBI isolates, with no synonymous nucleotide substitution. Western blotting and immunohistochemical analyses revealed that the amount of Imp expressed exceeded that of IdpA, in contrast to the case of a related phytoplasma-disease, western X-disease, for which the major membrane protein appears to be IdpA, not Imp. These results suggest that even phylogenetically close phytoplasmas express different types of major membrane proteins.
Introduction
The poinsettia (Euphorbia pulcherrima Wild. Klotz) is a native shrub of Mexico with brightly colored ‘flowers’ (bracts). Huge numbers of poinsettias are sold as ornamental plants during the Christmas season, amounting to approximately $240 million in 2005 in the United States (Floriculture and Nursery Crops Yearbook: http://www.ers.usda.gov) and $16 million in 2008 in Japan (The 84th Statistical Yearbook of Ministry of Agriculture Forestry and Fisheries: http://www.maff.go.jp/e/tokei/kikaku/nenji_e/index.html). Most commercially sold poinsettias are free branching, meaning they produce many axillary shoots and colored bracts and show reduced apical dominance. These characteristic features of free-branching poinsettias have been shown to be associated with poinsettia branch-inducing phytoplasma (PoiBI) (Lee et al., 1997), which decreases poinsettia height and increases branching. Thus, this particular bacterial infection increases the commercial value of these ornamental plants.
Phytoplasmas are pleomorphic bacteria of the class Mollicutes. As such, they lack cell walls and are obligate parasites of plants or insects. Phytoplasma infection is associated with devastating yield losses in many agriculturally important plant crops worldwide. Although the inability to culture phytoplasmas in vitro has hindered their biological characterization, the complete genome sequences of four phytoplasma strains [‘Candidatus Phytoplasma asteris’ strains OY-M and AY-WB (Oshima et al., 2004; Bai et al., 2006); ‘Candidatus Phytoplasma australiense’ strain AUSGY (Tran-Nguyen et al., 2008); and ‘Candidatus Phytoplasma mali’ strain AT (Kube et al., 2008)] have been determined. Analysis of these sequences has shown that phytoplasmas have lost many genes such as metabolic genes during their reductive evolution, presumably as an adaptation to living as intracellular parasites.
In contrast, phytoplasma genomes harbor many genes encoding membrane and secretory proteins. As phytoplasmas lack cell walls and are intracellular parasites, these proteins function in the cytoplasm of host cells, and are expected to have important functions in host–phytoplasma interactions. For example, they affect plant development as shown in TENGU, one of the secretory proteins of onion yellows phytoplasma (Hoshi et al., 2009). When tengu was expressed in Arabidopsis thaliana and Nicotiana benthamiana plants, these plants developed witches’ broom and dwarfism, which are typical symptoms of phytoplasma infection.
The majority of the phytoplasma surface is thought to be covered with membrane proteins known collectively as immunodominant membrane proteins (Imps) (Shen & Lin, 1993; Kakizawa et al., 2006a). Since they are principal components of the phytoplasma surface, Imps are often chosen as targets for the production of monoclonal and polyclonal antibodies used in phytoplasma detection (Clark et al., 1989; Yu et al., 1998; Berg et al., 1999; Blomquist et al., 2001; Barbara et al., 2002; Morton et al., 2003; Kakizawa et al., 2004, 2009).
Furthermore, since they are major proteins of the phytoplasma cell surface, Imps are predicted to play some important roles in phytoplasma–host interactions. The formation of a complex between antigenic membrane protein (Amp) of onion yellows phytoplasma and insect microfilaments has been correlated with the phytoplasma-transmitting capability of leafhoppers, suggesting that the interaction between Amp and insect microfilaments plays a role in phytoplasma transmissibility (Suzuki et al., 2006). Moreover, the Amp appears to have evolved under strong positive selection, indicating that it plays an important role in phytoplasma fitness (Kakizawa et al., 2006b, 2009).
Genes encoding Imps have been isolated from several phytoplasma groups, and have been classified into three types: (1) the specific Imp found in sweet potato witches’ broom (Yu et al., 1998), apple proliferation (Berg et al., 1999), European stone fruit yellows (Morton et al., 2003), pear decline (Morton et al., 2003), and peach yellow leaf roll (Morton et al., 2003) phytoplasmas; (2) immunodominant membrane protein A (IdpA), found in western X-disease (WX) phytoplasma (Blomquist et al., 2001); and (3) Amp, found in aster yellows (Barbara et al., 2002), clover phyllody (Barbara et al., 2002), and onion yellows (Kakizawa et al., 2004) phytoplasmas. These three types of proteins, Imp, IdpA, and Amp, share no amino acid sequence similarities and differ in their transmembrane structures. Several phytoplasma strains harbor genes encoding two types of these proteins and one of which is predominantly expressed [e.g. OY and WX encode imp, in addition to each major protein gene (Kakizawa et al., 2006a, 2009)]. Imp is conserved in many phytoplasmas, and might thus represent the ancestral Imp (Kakizawa et al., 2009).
PoiBI belongs to 16SrIII ribosomal group (Lee et al., 1998), which implies that the Imp of PoiBI might be IdpA, as it is in WX (Blomquist et al., 2001). Despite the commercial importance of PoiBI, its Imp has not been studied, and only a few of its genes have been cloned, such as those encoding the 16S rRNA gene-ITS-23S ribosomal RNA (rRNA) gene region, isoleucine tRNA, ribosomal protein L15, L22, protein translocase (secY), and methionine aminopeptidase (Martini et al., 2007; Lee et al., 2010). In the present study, we cloned both the imp and idpA genes from PoiBI, and analyzed Imp and IdpA protein expression in PoiBI-infected poinsettia cultivars. Contrary to expectation, the major membrane protein of PoiBI is Imp, and not IdpA. Moreover, as part of a detailed analysis of the biology and diversity of PoiBI, we examined the evolutionary implications of the Imp and IdpA protein sequences.
Materials and methods
PCR amplification of PoiBI
To detect phytoplasmas in poinsettia plants, total nucleic acid was extracted from the veinal tissues of 30 commercially available poinsettia cultivars (Table 1) using a DNeasy plant kit (Qiagen), according to the manufacturer's instructions. These nucleic acids were used as templates for ‘long and accurate’ PCR (LA-PCR) amplification of a 1.3-kb genome fragment expected to harbor the phytoplasma 16S rRNA gene. Reactions were performed in 25-μL mixtures containing 50–100 ng total nucleic acid, 0.5 μM each of primers SN910601 and SN910502 (Supporting Information, Table S1; Namba et al., 1993), 2.5 mM MgCl2, LA-PCR Buffer (Takara Bio), 0.8 U Takara LA Taq DNA polymerase (Takara Bio), and 400 μM each dNTP. An initial 2-min denaturation at 94 °C was followed by 35 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 60 °C, and extension for 90 s at 68 °C. In the final cycle, the 68 °C-extension step was extended to 7 min.
Detection of phytoplasmas in commercially available poinsettia cultivars
| Cultivar | Abbreviation | Detection of phytoplasma |
| Annette Hegg Dark Red | AHDR | + |
| Annette Hegg Lady | AHL | + |
| Annette Hegg Maxi | AHMaxi | + |
| Annette Hegg Pink | AHP | + |
| Annette Hegg Supreme | AHS | + |
| Arctic | Ar | + |
| Enduring Pink | EP | + |
| Enduring Red | ER | + |
| Enduring White | EW | + |
| Gutbier V-10 Amy | V-10 | + |
| Ice Punch | IP | + |
| Jester Jingle Bell | JJ | + |
| Jester Marble | JM | + |
| Jester Red | JR | + |
| Jingle Bells | JB | + |
| Monreale | M | + |
| Peterstar Marble | PSM | + |
| Peterstar Red | PSR | + |
| Premium Polar | PP | + |
| Premium Red | PR | + |
| Prestige Bright Red | PBR | + |
| Primero Jingle Bells | PJ | + |
| Silverstar Red | SR | + |
| Vision of Grandeur | VG | + |
| Winter Rose Pink | WP | + |
| Winter Rose White | WW | + |
| Annette Hegg Diva | AHD | − |
| Annette Hegg Marble | AHM | − |
| Eckespoint C-1 Red | C-1R | − |
| Flaming Sphere | FlS | − |
| Cultivar | Abbreviation | Detection of phytoplasma |
| Annette Hegg Dark Red | AHDR | + |
| Annette Hegg Lady | AHL | + |
| Annette Hegg Maxi | AHMaxi | + |
| Annette Hegg Pink | AHP | + |
| Annette Hegg Supreme | AHS | + |
| Arctic | Ar | + |
| Enduring Pink | EP | + |
| Enduring Red | ER | + |
| Enduring White | EW | + |
| Gutbier V-10 Amy | V-10 | + |
| Ice Punch | IP | + |
| Jester Jingle Bell | JJ | + |
| Jester Marble | JM | + |
| Jester Red | JR | + |
| Jingle Bells | JB | + |
| Monreale | M | + |
| Peterstar Marble | PSM | + |
| Peterstar Red | PSR | + |
| Premium Polar | PP | + |
| Premium Red | PR | + |
| Prestige Bright Red | PBR | + |
| Primero Jingle Bells | PJ | + |
| Silverstar Red | SR | + |
| Vision of Grandeur | VG | + |
| Winter Rose Pink | WP | + |
| Winter Rose White | WW | + |
| Annette Hegg Diva | AHD | − |
| Annette Hegg Marble | AHM | − |
| Eckespoint C-1 Red | C-1R | − |
| Flaming Sphere | FlS | − |
+ and − indicate cultivars in which PoiBI was detectable or not detectable, respectively, by PCR amplification of the phytoplasma 16S rRNA gene.
Detection of phytoplasmas in commercially available poinsettia cultivars
| Cultivar | Abbreviation | Detection of phytoplasma |
| Annette Hegg Dark Red | AHDR | + |
| Annette Hegg Lady | AHL | + |
| Annette Hegg Maxi | AHMaxi | + |
| Annette Hegg Pink | AHP | + |
| Annette Hegg Supreme | AHS | + |
| Arctic | Ar | + |
| Enduring Pink | EP | + |
| Enduring Red | ER | + |
| Enduring White | EW | + |
| Gutbier V-10 Amy | V-10 | + |
| Ice Punch | IP | + |
| Jester Jingle Bell | JJ | + |
| Jester Marble | JM | + |
| Jester Red | JR | + |
| Jingle Bells | JB | + |
| Monreale | M | + |
| Peterstar Marble | PSM | + |
| Peterstar Red | PSR | + |
| Premium Polar | PP | + |
| Premium Red | PR | + |
| Prestige Bright Red | PBR | + |
| Primero Jingle Bells | PJ | + |
| Silverstar Red | SR | + |
| Vision of Grandeur | VG | + |
| Winter Rose Pink | WP | + |
| Winter Rose White | WW | + |
| Annette Hegg Diva | AHD | − |
| Annette Hegg Marble | AHM | − |
| Eckespoint C-1 Red | C-1R | − |
| Flaming Sphere | FlS | − |
| Cultivar | Abbreviation | Detection of phytoplasma |
| Annette Hegg Dark Red | AHDR | + |
| Annette Hegg Lady | AHL | + |
| Annette Hegg Maxi | AHMaxi | + |
| Annette Hegg Pink | AHP | + |
| Annette Hegg Supreme | AHS | + |
| Arctic | Ar | + |
| Enduring Pink | EP | + |
| Enduring Red | ER | + |
| Enduring White | EW | + |
| Gutbier V-10 Amy | V-10 | + |
| Ice Punch | IP | + |
| Jester Jingle Bell | JJ | + |
| Jester Marble | JM | + |
| Jester Red | JR | + |
| Jingle Bells | JB | + |
| Monreale | M | + |
| Peterstar Marble | PSM | + |
| Peterstar Red | PSR | + |
| Premium Polar | PP | + |
| Premium Red | PR | + |
| Prestige Bright Red | PBR | + |
| Primero Jingle Bells | PJ | + |
| Silverstar Red | SR | + |
| Vision of Grandeur | VG | + |
| Winter Rose Pink | WP | + |
| Winter Rose White | WW | + |
| Annette Hegg Diva | AHD | − |
| Annette Hegg Marble | AHM | − |
| Eckespoint C-1 Red | C-1R | − |
| Flaming Sphere | FlS | − |
+ and − indicate cultivars in which PoiBI was detectable or not detectable, respectively, by PCR amplification of the phytoplasma 16S rRNA gene.
To clone the imp- and idpA-containing fragments of the PoiBI genome, DNA from the PoiBI-infected poinsettia cultivar ‘Primelo Jingle Bells’ was extracted and used as template for LA-PCR with three sets of primers (Fig. 1; Table S1). On the basis of the complete genomic sequence of OY-M (Oshima et al., 2004), we designed the primer pair PoiBI_imp-C01F/PssA-1 to amplify a 6.0-kb DNA fragment containing the imp gene. On the basis of a previously characterized WX DNA fragment (Liefting & Kirkpatrick, 2003), primer pair PoiBI_idpA-C1F/PoiBI_idpA-C2R was designed to amplify a 2.5-kb DNA fragment containing the idpA gene. Primer pair PoiBI_center-C1F/PoiBI_center-C2R was designed to amplify a 2.7-kb DNA fragment overlapping the sequence between the imp- and idpA-containing fragments. LA-PCRs were performed, as described above for amplification of the phytoplasma 16S rRNA gene, except that the annealing temperature was 53 °C and the extension time was 1 min kb−1. These amplified fragments were purified using ExoSAP-IT (Amersham Bioscience) and sequenced directly (primers shown in Table S1) using the dideoxynucleotide chain termination method on an automatic DNA sequencer (ABI PRISM 3130 Genetic Analyzer; Applied Biosystems), according to the manufacturer's instructions.
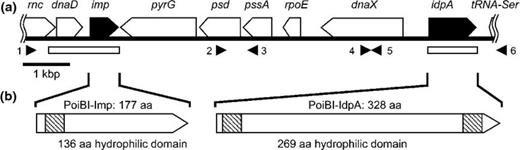
(a) Map of the 10.3-kbp DNA fragment amplified from PoiBI isolated from the poinsettia cv. ‘Primero Jingle Bells’. The fragment contains 10 genes encoding (in order) RNAse III (rnc; partial gene only), chromosome replication initiation protein (dnaD), immunodominant membrane protein (imp), CTP synthase (pyrG), phosphatidylserine decarboxylase (psd), phosphatidylserine synthase (pssA), DNA-directed RNA polymerase δ subunit (rpoE), DNA polymerase III (dnaX), immunodominant membrane protein A (idpA), and serine transfer RNA (tRNA-Ser; partial gene only). Filled arrow boxes indicate imp and idpA, and white arrow boxes indicate other genes. Filled arrowheads 1–6 under the map indicate the annealing positions of the primers PoiBI_imp-C01F, PoiBI_center-C1F, PssA-1, PoiBI_idpA-C1F, PoiBI_center-C2R, and PoiBI_idpA-C2R, respectively (see Table S1 for sequences), which were used for PCR amplification of DNA fragments. White boxes under the map indicate the completely sequenced regions of PoiBI isolates from 26 poinsettia cultivars. (b) Schematic representations of the Imp and IdpA proteins. Crosshatched boxes indicate transmembrane regions (TM) as predicted by the sosui program. aa, amino acids.
Thirty poinsettia cultivars were used as templates for amplification of the phytoplasma 16S rRNA gene. To investigate the sequence variability of PoiBI, we amplified and sequenced the imp- and idpA-containing genomic regions using the primer pairs PoiBI_imp-C02F/imp-R and idpAful-F/idpAful-R, respectively. These regions are shown in Fig. 1 as white boxes. The imp fragments were sequenced using primers PoiBI_imp-C02F, PoiBI_imp-C04F, imp-F, and imp-R. The idpA fragments were sequenced using primers idpAful-F, idpA532-F, idpA534-R, and idpAful-R. Primer sequences are shown in Table S1.
Sequence analysis and construction of a phylogenetic tree
The deduced amino acid sequences of Imp and IdpA from PoiBI and WX (Liefting & Kirkpatrick, 2003) were aligned using ClustalW (Thompson et al., 1994). The sequences were analyzed for the presence of putative transmembrane domains using the sosui program (ver. 1.11; http://bp.nuap.nagoya-u.ac.jp/sosui/sosui_submit.html; Hirokawa et al., 1998). The sequences were analyzed for the presence of secretion signal sequences using SignalP (ver. 3.0; http://www.cbs.dtu.dk/services/SignalP/) with the hidden Markov model (Bendtsen et al., 2004) and for protein localization using Psort (ver. 1; http://psort.hgc.jp/form.html; Horton et al., 2007).
Phylogenetic trees were constructed using mega (ver. 4; Tamura et al., 2007) with the neighbor-joining method and Poisson correction model.
Analysis of selection pressure
A comparison of synonymous and nonsynonymous nucleotide substitution rates is a useful approach for studying the mechanisms of DNA sequence evolution. The numbers of nonsynonymous and synonymous substitutions per site (dN and dS, respectively) and their ratio (dN/dS) are important indicators of selection pressure at the protein level; dN/dS values of <1, 1, and >1 imply stabilizing selection, neutral mutations, and diversifying positive selection, respectively. To examine positive selection pressure on dnaD, imp, and idpA of PoiBI, we used ClustalW (Thompson et al., 1994) to align the nucleotide sequences of dnaD, imp, and idpA of PoiBI and WX (Liefting & Kirkpatrick, 2003; GenBank Acc. No. AF533231). Alignments were adjusted manually, and dN/dS values were calculated as the overall average of the codon sites in each gene with Jukes-Cantor model of Nei-Gojobori method by mega (ver. 4; Tamura et al., 2007). A significant difference test for dN/dS was performed according to a previously described procedure (Messier & Stewart, 1997).
Expression of Imp and IdpA in Escherichia coli and antiserum production
For expression cloning, the entire imp gene from the Primelo Jingle Bells PoiBI isolate was PCR-amplified using primers impful-F and imp-R (Table S1), and a truncated form of the gene was PCR-amplified using primers impout-F and imp-R (Table S1). The truncated gene was designed to encode an Imp derivative lacking the N-terminal transmembrane region. Similarly, the entire idpA gene from the Primelo Jingle Bells PoiBI isolate was PCR-amplified using primers idpAful-F and idpAful-R, and a truncated form of the gene idpA gene was PCR-amplified using primers idpAcent-F and idpAcent-R. The truncated gene was designed to encode an IdpA derivative lacking both transmembrane regions and containing only the hydrophilic domain. In addition, idpA gene fragments encoding the N- and C-terminal halves of the IdpA hydrophilic domain were amplified using the primer pairs idpAcent-F/idpA534-R and idpA532-F/idpAcent-R, respectively.
A pET system (Novagen) was used to fuse histidine-tag (His-tag) and express the full-length and truncated Imp and IdpA proteins, as well as the IdpA hydrophilic fragments, in Escherichia coli. Each of the six PCR products described above was doubly digested with NdeI and XhoI and inserted into pET30a(+), thereby placing a His-tag at the C-terminus of the cloned fragments. The resulting constructs were transformed into E. coli BL21-CodonPlus (DE3)-RIL cells (Stratagene), and expression of the corresponding proteins was induced by the addition of isopropyl-β-d-thiogalactoside, according to the instructions provided by Stratagene. SDS-PAGE was performed to select the constructs expressing Imp or IdpA proteins of the proper size.
The His-tagged Imp and IdpA proteins were purified from the E. coli cell extracts by chromatography on a nickel NTA column (Qiagen), according to previously described procedures (Kakizawa et al., 2004). The purified proteins were used to immunize rabbits for preparation of antisera. The IgG fractions were purified from the crude sera with a Protein A Sepharose CL-4B (GE Healthcare, Piscataway, NJ).
Western blotting and immunohistochemical analysis
Western blotting was performed according to previously described procedures (Kakizawa et al., 2009) using anti-Imp and anti-IdpA IgG purified from immunized rabbits. Immunohistochemical analysis was performed according to a previously described method (Arashida et al., 2008) with some modifications. Stem tissues were excised from PoiBI-infected ‘Jester Red’ and uninfected ‘Flaming Sphere’ poinsettias, fixed, embedded in Paraplast Plus (Sherwood Medical), and cut into 10-μm thick sections using a microtome. Anti-Imp and anti-IdpA IgG were used with an alkaline phosphatase-mediated reporter system to detect Imp and IdpA proteins in each tissue. These tissues were observed by Axio Imager microscopy (Carl Zeiss).
Results
Detection of PoiBI in poinsettia plants
To detect PoiBI in poinsettia plants, we extracted total DNA from 30 commercially available poinsettia cultivars (Table 1) and amplified 1.3-kb DNA fragments containing the phytoplasma 16S rRNA gene by PCR. Of the 30 cultivars, all except ‘Annette Hegg Diva’, ‘Annette Hegg Marble’, ‘Eckespoint C-1 Red’, and ‘Flaming Sphere’ yielded fragments of the expected size (Table 1). Sequencing of these fragments confirmed that their DNA sequences were identical to that of the 16S rRNA gene of PoiBI (Lee et al., 1997; GenBank Acc. No. 190223), indicating that these 26 cultivars were infected with PoiBI.
Cloning of imp- and idpA-containing DNA fragments from PoiBI
Using total DNA isolated from the poinsettia cultivar ‘Primelo Jingle Bells’ as a template, we amplified a 6.0-kb DNA fragment containing the PoiBI imp gene, a 2.5-kb DNA fragment containing the PoiBI idpA gene, and a 3.3-kb DNA fragment between imp and idpA genes of the PoiBI DNA by LA-PCR (Fig. 1). Sequencing of these fragments yielded the complete DNA sequence of a 10-kb genomic region of PoiBI containing eight complete open reading frames and two partial open reading frames (Fig. 1). These genes (and their encoded proteins), listed in order, were rnc (RNAse III; partial gene only), dnaD (chromosome replication initiation protein), imp, pyrG (CTP synthase), psd (phosphatidylserine decarboxylase), pssA (phosphatidylserine synthase), rpoE (DNA-directed RNA polymerase δ subunit), dnaX (DNA polymerase III), idpA, and tRNA-Ser (serine transfer RNA; partial gene only). This gene structure is identical to that previously reported for WX strain (Liefting & Kirkpatrick, 2003; GenBank Acc. No. AF533231).
Cloning of PoiBI imp and idpA from multiple poinsettia cultivars
To investigate the sequence variability of PoiBI, we amplified and sequenced the dnaD-imp- and idpA-containing genomic regions of PoiBI using total DNA extracted from the 26 PoiBI-infected poinsettia cultivars. These two genomic regions are shown in Fig. 1 as white boxes. Both genomic fragments were successfully amplified from all 26 cultivars, and sequencing analysis confirmed that all 26 strains harbored dnaD, imp, and idpA. These two region sequences have been deposited in the GenBank database: AB636356-407. Analysis of the deduced amino acid sequences using sosui ver 1.11 (Hirokawa et al., 1998) yielded the predictions that Imp contains a transmembrane region in its N-terminal region and a hydrophilic domain, and that IdpA contains both N- and C-terminal transmembrane regions, as well as a central hydrophilic domain (Fig. 1b). These features are identical to those of other previously analyzed Imp and IdpA proteins (Kakizawa et al., 2006a, 2009).
Analysis of the Imp and IdpA sequences using the SignalP program with the hidden Markov model yielded the relatively high-probability predictions (0.885 and 0.824, respectively) that the two proteins have signal sequences. The signal sequence cleavage sites were predicted to lie between amino acid residues 48 and 49 for Imp, and between residues 35 and 36 for IdpA. Analysis of the Imp and IdpA sequences using the Psort program suggested with low probability (0.300) that Imp may be secreted from the bacterial cell and with high probability that IdpA is an integral membrane protein.
Sequence comparisons
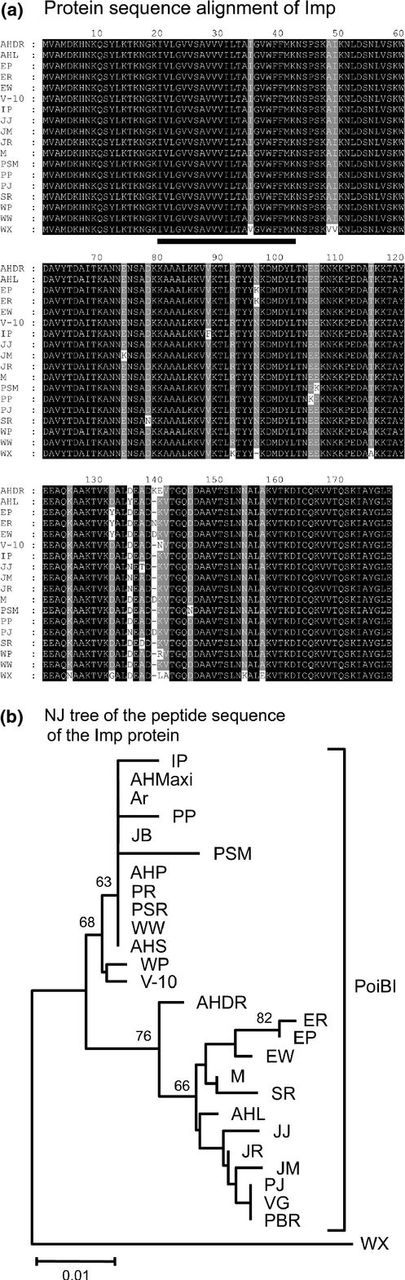
There were no silent substitutions in the PoiBI imp genes. Of the 26 PoiBI Imp amino acid sequences obtained from the 26 PoiBI-infected cultivars, those from ‘Annette Hegg Maxi’, ‘Annette Hegg Pink’, ‘Annette Hegg Supreme’, ‘Arctic’, ‘Jingle Bells’, ‘Premium Red’, and ‘Winter Rose White’ were 100% identical, and those from ‘Prestige Bright Red’, ‘Primero Jingle Bells’, and ‘Vision of Grandeur’ were identical. Therefore, in comparing the encoded Imp amino acid sequences, we used those from ‘Winter Rose White’ and ‘Primero Jingle Bells’, respectively, to represent these two groups of identical sequences. The resulting multiple alignment of these sequences and that of WX Imp is shown in Fig. 2a. Although variations in the PoiBI Imp sequences were noted at several positions, the sequence identity was overall very high. The lowest sequence identity score (97.2%) was obtained for the comparison of ‘Enduring Pink’ vs. ‘Jester Jingle Bells’, ‘Jester Marble’, and ‘Peterstar Marble’. A phylogenetic tree of the PoiBI Imp amino acid sequences is shown in Fig. 2b. In contrast to the diversity of imp genes, there was no difference in the sequences of the 16S rRNA gene, idpA, or dnaD genes from the 26 poinsettia cultivars.
Relationships among Imp protein sequences. (a) Alignment of amino acid sequences of Imp proteins from PoiBI and WX. (b) Phylogenetic neighbor-joining tree of Imp amino acid sequences. Numbers on the branches are bootstrap values (%) obtained for 1000 replicates. Abbreviations are given in Table 1.
The amino acid sequences deduced from PoiBI and WX imp, idpA, and dnaD are shown in Fig. S1. Among the PoiBI and WX Imp amino acid sequences, identity scores ranged from 92.6% to 93.8%, with a mean identity of 93.3%. The PoiBI and WX amino acid sequences of DnaD and IdpA had identity scores of 98.0% and 64.7%, respectively. The low sequence identity score of IdpA is partially attributed to insertions and deletions in the PoiBI IdpA sequence relative to that of WX IdpA.
Analysis of selection pressure
In a comparison of nucleotide sequences, the numbers of nonsynonymous and synonymous substitutions per site (dN and dS, respectively) and their ratio (dN/dS) are important indicators of selective pressure at the protein level. Calculated dN/dS values of <1, 1, and >1 imply stabilizing selection, neutral mutation, and diversifying positive selection, respectively. To investigate whether dnaD, imp, and idpA evolved under positive selection pressure, we determined the dN/dS value of the nucleotide sequences of these genes from PoiBI and WX. For dnaD, dN/dS was 0.444, which is < 1. For imp, dN/dS ranged from 1.278 to 1.556, but was not statistically significant (P = 0.3233–0.3716). In contrast, the dN/dS value for idpA was 1.500 with P = 0.0639, which is significant at the 10% level (Table 2).
The dN/dS scores and P values for comparisons of the idpA, imp, and dnaD gene sequences of PoiBI and WX
| dN/dS | P-values | |
| idpA | 1.50 | 0.0639 |
| imp | 1.28–1.56 | 0.3233–0.3716 |
| dnaD | 0.44 | 0.2991 |
| dN/dS | P-values | |
| idpA | 1.50 | 0.0639 |
| imp | 1.28–1.56 | 0.3233–0.3716 |
| dnaD | 0.44 | 0.2991 |
The dN/dS scores and P values for comparisons of the idpA, imp, and dnaD gene sequences of PoiBI and WX
| dN/dS | P-values | |
| idpA | 1.50 | 0.0639 |
| imp | 1.28–1.56 | 0.3233–0.3716 |
| dnaD | 0.44 | 0.2991 |
| dN/dS | P-values | |
| idpA | 1.50 | 0.0639 |
| imp | 1.28–1.56 | 0.3233–0.3716 |
| dnaD | 0.44 | 0.2991 |
Preparation of anti-PoiBI-Imp and anti-PoiBI-IdpA antibodies
To prepare materials for the production of anti-PoiBI-Imp and anti-PoiBI-IdpA antisera, we attempted to express His-tagged full-length and truncated forms of these proteins in E. coli. Attempts to express some proteins in E. coli were successful only for His-tagged full-length Imp and for His-tagged IdpA-N, which lacks the two transmembrane regions of IdpA, as well as half of the hydrophilic domain. These purified proteins were used to immunize rabbits. Use of the purified anti-Imp and anti-IdpA IgG in Western blots analysis of the Imp and IdpA hydrophilic domain proteins expressed in E. coli confirmed that the titers of the antibodies were similar (data not shown).
Western blot and immunohistochemical analyses
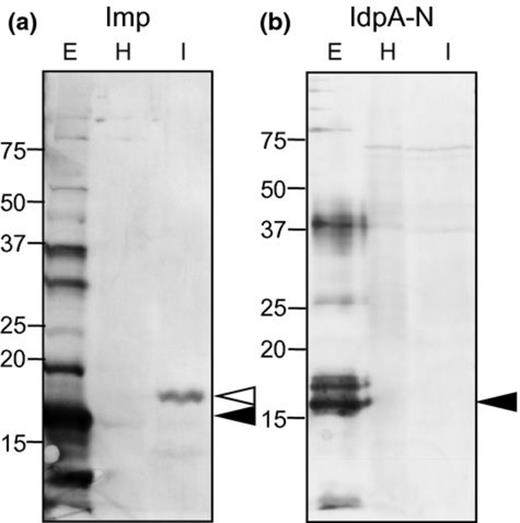
To confirm that Imp and IdpA are expressed in PoiBI-infected poinsettia, crude protein extracts were prepared from the PoiBI-infected cultivar ‘Jester Red’ (‘infected’ extract) and from the healthy control cultivar ‘Flaming Sphere’ (‘control’ extract). Western blot analysis of these samples using anti-Imp antibody revealed a distinct protein band in the infected extract, but not in the control extract (Fig. 3a). However, multiple attempts to detect IdpA using anti-IdpA antibody failed to detect IdpA in either the infected or control extracts (Fig. 3b).
Western blot analysis of extracts from PoiBI-infected and healthy poinsettias using antibodies against PoiBI-Imp and PoiBI-IdpA-N. H, extract from the noninfected (healthy) poinsettia cv. ‘Flaming Sphere’ (FlS); I, extract from the PoiBI-infected poinsettia cv. ‘Jester Red’ (JR); E, the purified Imp protein (a) and IdpA-N protein (b) expressed in Escherichia coli. Open and filled arrowheads indicate the major bands in each lane of the total protein extracted from the infected plant and purified proteins extracted from E. coli, respectively.
The Western blot analysis estimated the molecular mass of Imp in the ‘infected’ as 19.6 kDa, which is the calculated mass of full-length Imp. This result suggests that the signal sequence of Imp is not cleaved, and that Imp exists as a membrane protein in infected poinsettia plants. In contrast, the strongest Imp band detected in E. coli extracts was smaller by approximately 3 kDa, suggesting that the Imp signal sequence is cleaved when the recombinant protein is expressed in E. coli.
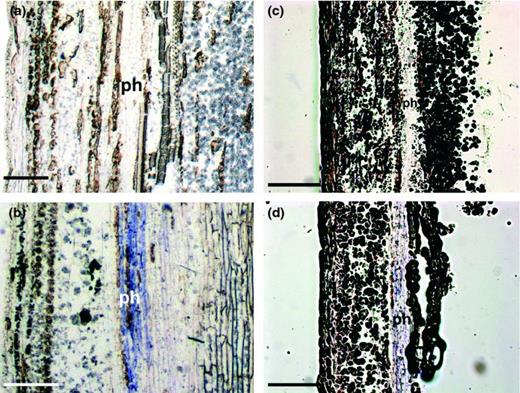
To investigate the localization of Imp and IdpA proteins in infected plants, we performed immunohistochemical analysis using anti-Imp and anti-IdpA antibodies. Both Imp and IdpA were specifically detected in the phloem of ‘Jester Red’ (‘infected’; Fig. 4b and d), but not in ‘Flaming Sphere’ (‘control’; Fig. 4a and c).
Immunohistochemical detection of Imp and IdpA protein expression in a vascular bundle of poinsettia stems. Anti-Imp antibody (a, b) and anti-IdpA antibody (c, d) were used to detect Imp and IdpA expression in stem longitudinal-sections of the PoiBI-free poinsettia cv. ‘Jester Red’ (JR) (a, c) and in the PoiBI-infected cv. ‘Flaming Sphere’ (FlS) (b, d). ph: phloem. Bars, 250 μm.
Discussion
Immunodominant membrane proteins of PoiBI
In this analysis of the imp and idpA genes of PoiBI, we confirmed the previously published assertions that these two genes are not homologous, and that imp is well conserved over a wide range of phytoplasma strains (Kakizawa et al., 2009). Although the Imp of WX has been previously reported to be IdpA (Blomquist et al., 2001), the identity of the Imp of PoiBI was not studied. In the present study, we were able to detect the expression of Imp in PoiBI-infected poinsettia plants using both immunohistochemical and Western blot analyses (Fig. 3a and b Fig. 4). In contrast, although we were able to detect expression of IdpA in PoiBI-infected plants immunohistochemically, we were not able to detect it by Western blotting, probably because immunohistochemical analysis is generally the more sensitive technique (Jiang et al., 1988; Friedlander et al., 1989; Gala et al., 1994). Our findings suggest that Imp is expressed at a higher level than IdpA in PoiBI.
In comparing PoiBI strains from 26 different poinsettia cultivars, we found no variation in their idpA, dnaD, or 16S rRNA genes. On the other hand, imp did exhibit some variation. All of the nucleotide substitutions in PoiBI imp resulted in amino acid changes; that is, no silent mutations were observed, suggesting that imp is prone to mutation. Although adaptive evolution of imp was not detected (Table 2), strong positive selection has been reported for the imp genes of AY-group phytoplasmas, indicating that Imp plays an important role in phytoplasma fitness (Kakizawa et al., 2006b, 2009). The imp gene of PoiBI might also play a crucial role correlated to the accumulation of amino acid substitutions.
AY-group phytoplasmas express approximately ten times as much Amp as Imp, indicating that the Imp in this group is Amp (Kakizawa et al., 2009). Among phytoplasma strains, amp gene sequences exhibit much more variation than imp gene sequences, and are subject to strong positive selection pressure (Kakizawa et al., 2009). In PoiBI, Imp was expressed at a higher level than IdpA (Fig. 3a and b), suggesting that the major membrane protein of PoiBI is Imp rather than IdpA, even though the Imp in closely related WX is IdpA (Blomquist et al., 2001). Therefore, the genes encoding Imps appear to differ among even closely related phytoplasma strains. Further analyses of imp and idpA sequences and gene expression among many strains of 16SrIII ribosomal group phytoplasmas, which include PoiBI and WX should yield more information about the diversity of Imps.
Comparison of PoiBI and WX
The average nucleotide sequence identity between PoiBI imp genes and WX imp in our study was 97.2%, whereas that between PoiBI and WX idpA was 77.7%. Nonsynonymous substitutions outnumbered synonymous substitutions for both genes from the two strains, and dN/dS was > 1 for both comparisons (Table 2). The high value of dN/dS for idpA resulted solely from the differences between WX and PoiBI idpA, as there was no variation among the 26 PoiBI sequences. As IdpA is the Imp of WX as reported (Blomquist et al., 2001), amino acid substitutions in WX IdpA should affect the properties of the WX cell surface and subsequently increase the evolutionary fitness of WX. These results suggest that IdpA has an important role in host–phytoplasma interactions, particularly in WX. Further sequence comparisons of Imp or IdpA among several strains of WX would reveal the functional importance of Imps in 16SrIII ribosomal group phytoplasmas.
Detection of PoiBI in poinsettia cultivars
Most of the 30 poinsettia cultivars examined in this study were infected with PoiBI, as shown by PCR amplification of the phytoplasma 16S rRNA gene, but phytoplasma infection could not be detected in four of the cultivars: ‘Flaming Sphere’, ‘Annette Hegg Marble’, ‘Annette Hegg Diva’, and ‘Eckespoint C-1 Red’. ‘Eckespoint C-1 Red’ was previously reported to be phytoplasma-free, along with ‘Eckespoint C-1 White’ (Dole & Wilkins, 1991), in agreement with our results. However, we cannot exclude the possibility that PCR failures resulted in false negatives for some or all of these cultivars. PCR failures could have arisen if the level of PoiBI accumulation was very low, perhaps as a result of the particular cultivar characteristics, growth stage, or growth conditions, or if the cultivar(s) contained PCR inhibitory compounds. Alternatively, the possibility of the sequence variability in the PCR primer binding region cannot be excluded. It is possible that nested-PCR using 16SrIII group-specific primers, instead of the single PCR using the primers in this study, might yield amplification products. However, we extracted the template DNA for all samples from the poinsettia leaf midribs, where the concentration of phytoplasma cells is expected to be high, and we followed the same extraction protocol for all poinsettia cultivars. To eliminate the influence of PCR inhibitory compounds, we used DNA diluted by tenth and hundredth as a template for PCR amplification. However, we could not yield fragments from all of four cultivars (data not shown). Moreover, phenotypically, these four cultivars are taller and had less branching than PoiBI-infected poinsettias (Fig. S2). These features were similar to those of the healthy poinsettia. Therefore, we conclude that in addition to ‘Eckespoint C-1 Red’ and ‘Eckespoint C-1 White’, several other commercially available poinsettias, that is, ‘Flaming Sphere’, ‘Annette Hegg Marble’, and ‘Annette Hegg Diva’, are free of phytoplasma infection.
Evolution of immunodominant membrane proteins
The conservation of Imp sequences among many groups of phytoplasmas has led to the suggestion that Imp represents an ancestral type of Imp (Kakizawa et al., ). This proposal suggests that PoiBI has retained Imp as its major membrane protein, and that the expression level of IdpA had increased during the evolution of WX, causing IdpA to become the Imp of WX. It is known that WX is transmitted predominantly by Colladonus montanus (Kirkpatrick et al., 1987), whereas it has been assumed that PoiBI is transmitted only by grafting. In addition, phytoplasma genomes show high plasticity. For example, when phytoplasma was maintained by grafting or tissue culture, its insect-transmissibility was easily lost and genes involved in the phytoplasma-insect interactions were mutated (Oshima et al., 2001; Ishii et al.,2009a, b). Based on this difference of modes of transmission between WX and PoiBI and on the genome plasticity of phytoplasmas, the membrane proteins of the two phytoplasmas may have evolved in different ways. Further analyses of the diversity and functions of Imps are expected to reveal the evolution and biology of phytoplasmas.
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research (21248004) and the Funding Program for Next Generation World-Leading Researchers of Japan Society for the Promotion Science, and also by the Program for Promotion of Basic Research Activities for Innovative Bioscience of Bio-oriented Technology Research Advancement Institution.
References
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Amino acid sequence alignment of IdpA (a), and DnaD (chromosome replication initiation protein) (b) from WX and PoiBI isolated from the poinsettia cv. ‘Primero Jingle Bells’.
Fig. S2. Phenotypes of restricted- and free-branching poinsettia plants.
Table S1. Sequences of PCR primers used in this study.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Author notes
Editor: Herman Bothe