-
PDF
- Split View
-
Views
-
Cite
Cite
Donald M. Gardiner, Barbara J. Howlett, Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus, FEMS Microbiology Letters, Volume 248, Issue 2, July 2005, Pages 241–248, https://doi.org/10.1016/j.femsle.2005.05.046
Close - Share Icon Share
Abstract
Gliotoxin is a secondary metabolite produced by several fungi including the opportunistic animal pathogen Aspergillus fumigatus. It is a member of the epipolythiodioxopiperazine (ETP) class of toxins characterised by a disulphide bridged cyclic dipeptide. A putative cluster of 12 genes involved in gliotoxin biosynthesis has been identified in A. fumigatus by a comparative genomics approach based on homology to genes from the sirodesmin (another ETP) biosynthetic gene cluster of Leptosphaeria maculans. The physical limits of the cluster in A. fumigatus have been defined by bioinformatics and by identifying the genes that are co-regulated and whose timing of expression correlates with the production of gliotoxin in culture.
1 Introduction
Gliotoxin is a secondary metabolite produced by several filamentous fungi including Aspergillus fumigatus, some Penicillium spp. and a few clinical isolates of Candida albicans, which are opportunistic animal pathogens, as well as mycoparasitic Trichoderma spp. that are used in biocontrol of plant pathogens [[,[]. Gliotoxin is an epipolythiodioxopiperazine (ETP), a class of cyclic dipeptides characterised by the presence of an internal disulphide bridge [[]. There are two reported modes of toxicity. Firstly, they act as redox active toxins generating reactive oxygen species by cycling between their oxidised (disulfide) and reduced (dithiol) forms [[]. Secondly they form mixed disulphides with proteins that have accessible thiol groups [[]. Additionally ETP molecules are actively concentrated within the target cell which enhances their toxicity [[].
Gliotoxin has immunosuppressive properties [[]. A. fumigatus is the major cause of invasive aspergillosis, a complication in leukemia sufferers, organ transplant recipients and HIV-AIDS patients and gliotoxin is indirectly implicated in these activities [[]. Additionally in vitro experiments with mammalian cell lines show that gliotoxin causes both apoptotic and necrotic cell death [[0,[1].
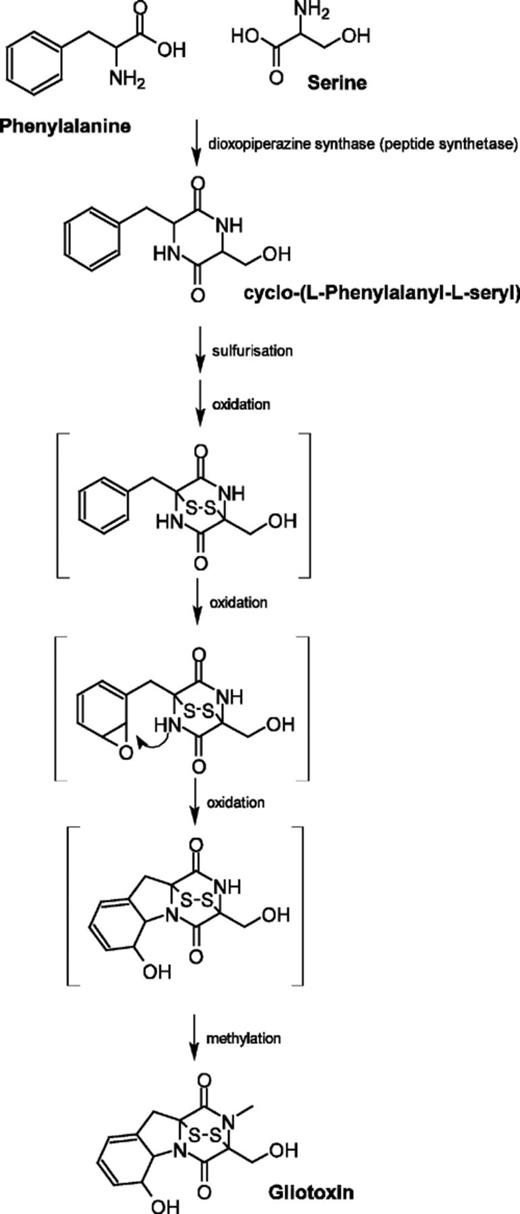
In spite of the important biological effects of gliotoxin, genes encoding enzymes responsible for its biosynthesis have not been identified. The first step is likely to involve condensation of serine and phenylalanine predicted to be catalysed by a non-ribosomal peptide synthetase. Subsequently, as for sirodesmin (another ETP), a series of oxidations, sulfurisation, and methylation are predicted (Fig. 1).
Proposed pathway for gliotoxin biosynthesis. The only known intermediate is cyclo l-phenylalanyl-l-serine. All other compounds are predicted (bracketed) and the order of reactions may not be correct.
Recently, we reported the first identification of a cluster of biosynthetic genes for an ETP, sirodesmin, from the phytopathogenic fungus, Leptosphaeria maculans[[2]. This cluster contained 18 genes and roles for many of them in sirodesmin production were deduced. Disruption of one gene (a non-ribosomal peptide synthetase) resulted in a mutant unable to make sirodesmin. Since complete genome sequences are now available for several other fungi, these sequences were searched for linked genes (non-ribosomal peptide synthetase and thioredoxin reductase) predicted to be essential for ETP production. A gene cluster including these two genes as well as other homologs of the sirodesmin cluster was detected in the A. fumigatus genome (http://www.tigr.org/). Furthermore, this cluster was not present in A. nidulans, which does not make gliotoxin. The putative transport protein present in this cluster has also been shown to provide resistance to gliotoxin when expressed in an L. maculans mutant sensitive to gliotoxin [[3].
In this paper, we compare the arrangement and sequence of genes in the putative gliotoxin biosynthetic cluster in A. fumigatus to those in the sirodesmin biosynthetic cluster. We also describe the regulation of genes in the putative gliotoxin biosynthetic cluster and those flanking it.
2 Materials and methods
2.1 Fungal culturing
Aspergillus fumigatus isolate Af293 was grown on 10% (v/v) Campbells V8 juice agar plates and Czapek Dox liquid media at 37°C. Conidiophores were harvested by flooding plates with sterile 0.5% (v/v in water) Tween 20. To analyse gene expression over time, cultures (107 spores in 15 mL) were harvested at 16, 24, 48 and 96 h after inoculation of the cultures. For the 8 h culture, 108 spores were used for culture inoculation.
2.2 Quantitative RT-PCR analysis
The expression of 12 genes from the putative gliotoxin biosynthetic gene cluster and five flanking genes were analysed using quantitative RT-PCR (qRT-PCR) using methods described in Gardiner et al. [[2]. RNA was purified using the RNeasy Plant Mini kit (QIAgen, Germany) and DNaseI treated (Invitrogen) prior to oligo-dT primed reverse transcription with SuperScript III (Invitrogen). Quantitative RT-PCR was performed using Rotor-Gene 3000 equipment (Corbett Research, Australia) and QuantiTect™ SYBR? Green PCR kit (QIAgen). A standard curve of amplification efficiency of each gene was generated from purified RT-PCR products across a five orders of magnitude dilution series (10−4–10−8 dilution) in triplicate. Samples were analysed in triplicate from a fivefold dilution of the original RT products. Diluted RT product (1 μL) was added to 19 μL of PCR mix and subjected to 45 cycles of PCR (30 s at each of 94, 60–64 and 72°C, with the annealing temperature optimised for particular primers). The amplified product was detected every cycle at the end of the 72°C step. Melt curve analysis after the cycling confirmed the absence of non-specific products in the reaction. The fluorescence threshold (Ct) values were determined for standards and samples using the Rotor-Gene 5 software and typically in the range between 15 and 25 PCR cycles. Data were processed according to Muller et al. [[4]. All values are relative to actin in that sample (A. fumigatus Locus ID Afu6g04740). To enable comparison between genes, the data are normalised to the relative expression at 24 h (i.e., the value of expression at 24 h is given a value of one). Primer sequences are given in Table 1.
Primers used in quantitative RT-PCR
| Primer namea | Sequence (5′-3′) |
| Afactinf | CTCCAGCTTGGAGAAGTCCT |
| cgrid Afactinr | CGGACGTCGACATCACACTT |
| AN7527.2hf | TCATTATCGAGCACCACCAA |
| AN7527.2hr | ACAGGCCATGGATGAGAAAG |
| AN7529.2hf | GACGGGCTGTACGACAAAGT |
| AN7529.2hr | GTCACCTCCCGACTCACAAT |
| CPPS2hf | CAGCTTCTTCCACCTTGGAG |
| CPPS2hr | TATCTGGCGCATCTCTTGTG |
| GliZf | ACGACGATGAGGAATCGAAC |
| GliZr | TCCAGAAAAGGGAGTCGTTG |
| GliIf | AGGCCATCCTCGTGTGTAAC |
| GliIr | GCCGAGGTCTTTGCTGATAC |
| GliJf | CTCTGATCGACGGCCATAAT |
| GliJr | TCGAGCTGTTGGAGTGTCTG |
| GliPf | AAACCCCTGTGAATGCAGAC |
| GliPr | CCCCTTGAGATGAAAGGTGA |
| GliCf | ATTGACCGGGATGACACATT |
| GliCr | ACCGTCGAGGATTGTATTGC |
| GliMf | CGATCTGTACCCCAACGAGT |
| GliMr | TTCTGGAACTTTGCCAGCTT |
| GliGf | GAAACTGCGCAGCAACATTA |
| GliGr | TTGGCCATTTCTCAAACTCC |
| GliKf | CTCACGGCATACAGCGACTA |
| GliKr | ATAATCCAACCGAGCCACTG |
| GliAf | TTTGCGATCAACGAACTCTG |
| GliAr | CCCTTGACGGACTGGAAGTA |
| GliNf | GCAAGAGGTGCAAGAGAAGG |
| GliNr | GGATCGGATCAAAGTCCTCA |
| GliFf | GGGGGCCGATAATATCAACT |
| GliFr | AAGATGGCCAATCCACCATA |
| GliTf | ACTCCACCATCCAGTTCCAG |
| GliTr | TCCGAGTATCCCTCGATGTC |
| AN2610.2hf | TGCCACATTCAGGGATTGTA |
| AN2610.2hr | GAAAGTTGCGGAAAAGTTCG |
| AN2611.2hf | AGCTACTCGTCGACCTGGAA |
| AN2611.2hr | TGGCGAGCTCTAGTCCATTT |
| Primer namea | Sequence (5′-3′) |
| Afactinf | CTCCAGCTTGGAGAAGTCCT |
| cgrid Afactinr | CGGACGTCGACATCACACTT |
| AN7527.2hf | TCATTATCGAGCACCACCAA |
| AN7527.2hr | ACAGGCCATGGATGAGAAAG |
| AN7529.2hf | GACGGGCTGTACGACAAAGT |
| AN7529.2hr | GTCACCTCCCGACTCACAAT |
| CPPS2hf | CAGCTTCTTCCACCTTGGAG |
| CPPS2hr | TATCTGGCGCATCTCTTGTG |
| GliZf | ACGACGATGAGGAATCGAAC |
| GliZr | TCCAGAAAAGGGAGTCGTTG |
| GliIf | AGGCCATCCTCGTGTGTAAC |
| GliIr | GCCGAGGTCTTTGCTGATAC |
| GliJf | CTCTGATCGACGGCCATAAT |
| GliJr | TCGAGCTGTTGGAGTGTCTG |
| GliPf | AAACCCCTGTGAATGCAGAC |
| GliPr | CCCCTTGAGATGAAAGGTGA |
| GliCf | ATTGACCGGGATGACACATT |
| GliCr | ACCGTCGAGGATTGTATTGC |
| GliMf | CGATCTGTACCCCAACGAGT |
| GliMr | TTCTGGAACTTTGCCAGCTT |
| GliGf | GAAACTGCGCAGCAACATTA |
| GliGr | TTGGCCATTTCTCAAACTCC |
| GliKf | CTCACGGCATACAGCGACTA |
| GliKr | ATAATCCAACCGAGCCACTG |
| GliAf | TTTGCGATCAACGAACTCTG |
| GliAr | CCCTTGACGGACTGGAAGTA |
| GliNf | GCAAGAGGTGCAAGAGAAGG |
| GliNr | GGATCGGATCAAAGTCCTCA |
| GliFf | GGGGGCCGATAATATCAACT |
| GliFr | AAGATGGCCAATCCACCATA |
| GliTf | ACTCCACCATCCAGTTCCAG |
| GliTr | TCCGAGTATCCCTCGATGTC |
| AN2610.2hf | TGCCACATTCAGGGATTGTA |
| AN2610.2hr | GAAAGTTGCGGAAAAGTTCG |
| AN2611.2hf | AGCTACTCGTCGACCTGGAA |
| AN2611.2hr | TGGCGAGCTCTAGTCCATTT |
Primers used in quantitative RT-PCR
| Primer namea | Sequence (5′-3′) |
| Afactinf | CTCCAGCTTGGAGAAGTCCT |
| cgrid Afactinr | CGGACGTCGACATCACACTT |
| AN7527.2hf | TCATTATCGAGCACCACCAA |
| AN7527.2hr | ACAGGCCATGGATGAGAAAG |
| AN7529.2hf | GACGGGCTGTACGACAAAGT |
| AN7529.2hr | GTCACCTCCCGACTCACAAT |
| CPPS2hf | CAGCTTCTTCCACCTTGGAG |
| CPPS2hr | TATCTGGCGCATCTCTTGTG |
| GliZf | ACGACGATGAGGAATCGAAC |
| GliZr | TCCAGAAAAGGGAGTCGTTG |
| GliIf | AGGCCATCCTCGTGTGTAAC |
| GliIr | GCCGAGGTCTTTGCTGATAC |
| GliJf | CTCTGATCGACGGCCATAAT |
| GliJr | TCGAGCTGTTGGAGTGTCTG |
| GliPf | AAACCCCTGTGAATGCAGAC |
| GliPr | CCCCTTGAGATGAAAGGTGA |
| GliCf | ATTGACCGGGATGACACATT |
| GliCr | ACCGTCGAGGATTGTATTGC |
| GliMf | CGATCTGTACCCCAACGAGT |
| GliMr | TTCTGGAACTTTGCCAGCTT |
| GliGf | GAAACTGCGCAGCAACATTA |
| GliGr | TTGGCCATTTCTCAAACTCC |
| GliKf | CTCACGGCATACAGCGACTA |
| GliKr | ATAATCCAACCGAGCCACTG |
| GliAf | TTTGCGATCAACGAACTCTG |
| GliAr | CCCTTGACGGACTGGAAGTA |
| GliNf | GCAAGAGGTGCAAGAGAAGG |
| GliNr | GGATCGGATCAAAGTCCTCA |
| GliFf | GGGGGCCGATAATATCAACT |
| GliFr | AAGATGGCCAATCCACCATA |
| GliTf | ACTCCACCATCCAGTTCCAG |
| GliTr | TCCGAGTATCCCTCGATGTC |
| AN2610.2hf | TGCCACATTCAGGGATTGTA |
| AN2610.2hr | GAAAGTTGCGGAAAAGTTCG |
| AN2611.2hf | AGCTACTCGTCGACCTGGAA |
| AN2611.2hr | TGGCGAGCTCTAGTCCATTT |
| Primer namea | Sequence (5′-3′) |
| Afactinf | CTCCAGCTTGGAGAAGTCCT |
| cgrid Afactinr | CGGACGTCGACATCACACTT |
| AN7527.2hf | TCATTATCGAGCACCACCAA |
| AN7527.2hr | ACAGGCCATGGATGAGAAAG |
| AN7529.2hf | GACGGGCTGTACGACAAAGT |
| AN7529.2hr | GTCACCTCCCGACTCACAAT |
| CPPS2hf | CAGCTTCTTCCACCTTGGAG |
| CPPS2hr | TATCTGGCGCATCTCTTGTG |
| GliZf | ACGACGATGAGGAATCGAAC |
| GliZr | TCCAGAAAAGGGAGTCGTTG |
| GliIf | AGGCCATCCTCGTGTGTAAC |
| GliIr | GCCGAGGTCTTTGCTGATAC |
| GliJf | CTCTGATCGACGGCCATAAT |
| GliJr | TCGAGCTGTTGGAGTGTCTG |
| GliPf | AAACCCCTGTGAATGCAGAC |
| GliPr | CCCCTTGAGATGAAAGGTGA |
| GliCf | ATTGACCGGGATGACACATT |
| GliCr | ACCGTCGAGGATTGTATTGC |
| GliMf | CGATCTGTACCCCAACGAGT |
| GliMr | TTCTGGAACTTTGCCAGCTT |
| GliGf | GAAACTGCGCAGCAACATTA |
| GliGr | TTGGCCATTTCTCAAACTCC |
| GliKf | CTCACGGCATACAGCGACTA |
| GliKr | ATAATCCAACCGAGCCACTG |
| GliAf | TTTGCGATCAACGAACTCTG |
| GliAr | CCCTTGACGGACTGGAAGTA |
| GliNf | GCAAGAGGTGCAAGAGAAGG |
| GliNr | GGATCGGATCAAAGTCCTCA |
| GliFf | GGGGGCCGATAATATCAACT |
| GliFr | AAGATGGCCAATCCACCATA |
| GliTf | ACTCCACCATCCAGTTCCAG |
| GliTr | TCCGAGTATCCCTCGATGTC |
| AN2610.2hf | TGCCACATTCAGGGATTGTA |
| AN2610.2hr | GAAAGTTGCGGAAAAGTTCG |
| AN2611.2hf | AGCTACTCGTCGACCTGGAA |
| AN2611.2hr | TGGCGAGCTCTAGTCCATTT |
2.3 RP-HPLC analysis
Duplicate culture filtrates (4 mL) of A. fumigatus were extracted as described previously [[2] in the presence of sirodesmin (50 μg) as an internal control. RP-HPLC was performed as described previously except that a Phenomenex Synergi 4 μm particle size, 4.6 i.d. × 250 mm C18(2) column was used [[2].
3 Results
3.1 Comparison of the putative gliotoxin biosynthetic gene cluster to the sirodesmin biosynthetic gene cluster
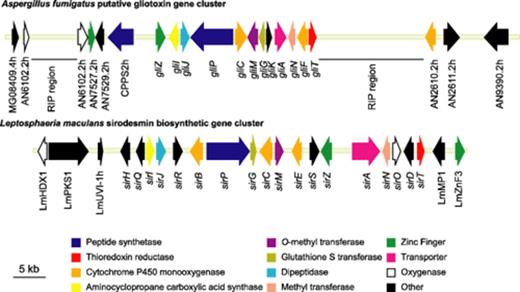
Two similar gene clusters, one from L. maculans encoding enzymes in sirodesmin biosynthetic gene cluster, and the putative gliotoxin biosynthetic cluster in A. fumigatus are shown in Fig. 2. Analysis of the sequence similarity shared between the gene products encoded by the putative gliotoxin cluster and those in the sirodesmin cluster is summarised in Table 2. Eight genes are highly conserved between the two clusters (e-value less than e−60) and these are deemed as core ETP moiety genes. The peptide synthetase (GliP) had identical domain architecture to that of SirP, containing two complete modules and an additional phosphopantetheine attachment site (Supplementary figure 1). Two other genes, encoding the transcriptional regulators (sirZ and gliZ) and the transporters (sirA and gliA) are also likely to be functional equivalents. The transporters are from different classes; SirA is an ABC transporter, whereas the gliotoxin transporter, GliA, is a member of the major facilitator superfamily of transporters. The two genes that had no obvious homologs in the sirodesmin biosynthetic gene cluster had best matches to a cytochrome P450 monooxygenase (GliF) and a hypothetical protein (GliK). A GliK homologue was present in an ETP-like gene cluster identified from the rice blast fungus, Magnaporthe grisea[[2]. It appeared to be fused to another gene, but this maybe due to inaccurate annotation of this region of M. grisea.
Comparison of the putative gliotoxin and sirodesmin biosynthetic gene clusters from Aspergillus fumigatus and Leptosphaeria maculans, respectively. The “other” category contains genes encoding cytochrome P450 monooxygenases (GliF, SirB, SirE), a prenyl transferase (SirD), an acetyl transferase (SirH), epimerases (SirQ, SirR, SirS), an oxidoreductase (SirO) and a hypothetical protein (GliK). A total of 68 kb of L. maculans and 81.5 kb of sequence of A. fumigatus, respectively is represented. The RIP regions in A. fumigatus (repeat induced point mutation) are AT-rich compared to surrounding regions and contain remnants of retroelements. The region encoding the gliotoxin gene cluster has been submitted to GenBank (Accession AY838877). Genes outside of the gliotoxin cluster are named by the closest homologue (e.g., AN2610.2h).
Homologies between the A. fumigatus putative gliotoxin biosynthetic gene cluster and the L. maculans sirodesmin biosynthetic gene cluster
| Gene namea | Sirodesmin cluster homologue | BLASTX match | e-value | Encoded product/commentsb |
| − | − | MG08409.4 | 7e−61 | Endoglucanase |
| − | − | AN6102.2 | 4e−84 | Quinone oxidoreductase |
| − | − | AN7527.2 | 5e−91 | Zinc finger transcription factor |
| − | − | AN7529.2 | 5e−150 | Peptidase (Afu6g09600) |
| − | − | CPPS2, Claviceps purpurea (CAD28788) | 2e−109 | Mono-modular peptide synthetase (Afu6g09610) |
| gliZ | sirZ | SirZ | 5e−10 | Zinc finger transcriptional regulator |
| gliI | sirI | SirI | 2e−61 | 1-aminocyclopropane-1-carboxylic acid synthase (ACCS). Core ETP biosynthesis. |
| gliJ | sirJ | MG07739.4 | 2e−92 | Dipeptidase. Core ETP biosynthesis. |
| gliP | sirP | SirP | 0 | Two module non-ribosomal peptide synthetase. Core ETP biosynthesis. |
| gliC | sirC | MG07743.4 | 3e−92 | Cytochrome P450 monooxygenase sharing similarity with SirC. Core ETP biosynthesis. |
| gliM | sirM | SirM | 1e−80 | O-Methyl transferase. Core ETP biosynthesis. |
| gliG | sirG | Alternaria alternata GST (AAR98813) | 3e−78 | Glutathione S-transferase. Core ETP biosynthesis. |
| gliK | none | MG07739.4 | 2e−35 | Hypothetical protein, no matches in sirodesmin biosynthetic gene cluster. |
| gliA | none | FG04188.1 | e−103 | Major facilitator type transporter. Functional equivalent of SirA. |
| gliN | sirN | SirN | 2e−75 | Methyl transferase. Core ETP biosynthesis. |
| gliF | none | AN1598.2 | 9e−54 | Cytochrome P450 monooxygenase. Not necessarily functional equivalents of SirB or SirE |
| gliT | sirT | AN3963.2 | 1e−70 | Thioredoxin reductase. Core ETP biosynthesis. |
| AN2610.2 | 0 | Cytochrome P450 monooxygenase trichothecene C15 hydroxylase (Tri11) (Afu6g09760) | ||
| − | AN2611.2 | e−117 | Domain matches to terpene cyclase and transisoprenyl pyrophosphate synthase. Probably involved in terpenoid (secondary) metabolism. (Afu6g09770) | |
| − | AN9390.2 | 3e−95 | Chitinase (Afu6g09780) |
| Gene namea | Sirodesmin cluster homologue | BLASTX match | e-value | Encoded product/commentsb |
| − | − | MG08409.4 | 7e−61 | Endoglucanase |
| − | − | AN6102.2 | 4e−84 | Quinone oxidoreductase |
| − | − | AN7527.2 | 5e−91 | Zinc finger transcription factor |
| − | − | AN7529.2 | 5e−150 | Peptidase (Afu6g09600) |
| − | − | CPPS2, Claviceps purpurea (CAD28788) | 2e−109 | Mono-modular peptide synthetase (Afu6g09610) |
| gliZ | sirZ | SirZ | 5e−10 | Zinc finger transcriptional regulator |
| gliI | sirI | SirI | 2e−61 | 1-aminocyclopropane-1-carboxylic acid synthase (ACCS). Core ETP biosynthesis. |
| gliJ | sirJ | MG07739.4 | 2e−92 | Dipeptidase. Core ETP biosynthesis. |
| gliP | sirP | SirP | 0 | Two module non-ribosomal peptide synthetase. Core ETP biosynthesis. |
| gliC | sirC | MG07743.4 | 3e−92 | Cytochrome P450 monooxygenase sharing similarity with SirC. Core ETP biosynthesis. |
| gliM | sirM | SirM | 1e−80 | O-Methyl transferase. Core ETP biosynthesis. |
| gliG | sirG | Alternaria alternata GST (AAR98813) | 3e−78 | Glutathione S-transferase. Core ETP biosynthesis. |
| gliK | none | MG07739.4 | 2e−35 | Hypothetical protein, no matches in sirodesmin biosynthetic gene cluster. |
| gliA | none | FG04188.1 | e−103 | Major facilitator type transporter. Functional equivalent of SirA. |
| gliN | sirN | SirN | 2e−75 | Methyl transferase. Core ETP biosynthesis. |
| gliF | none | AN1598.2 | 9e−54 | Cytochrome P450 monooxygenase. Not necessarily functional equivalents of SirB or SirE |
| gliT | sirT | AN3963.2 | 1e−70 | Thioredoxin reductase. Core ETP biosynthesis. |
| AN2610.2 | 0 | Cytochrome P450 monooxygenase trichothecene C15 hydroxylase (Tri11) (Afu6g09760) | ||
| − | AN2611.2 | e−117 | Domain matches to terpene cyclase and transisoprenyl pyrophosphate synthase. Probably involved in terpenoid (secondary) metabolism. (Afu6g09770) | |
| − | AN9390.2 | 3e−95 | Chitinase (Afu6g09780) |
Not all gli gene products had a best match to the equivalent from L. maculans. Some showed higher sequence similarity to products encoded by the M. grisea cluster [[2]. Four (GliA, GliF, GliG and GliT) showed higher sequence similarity to gene products from other species, but all these are members of multigene families.
aGene nomenclature is based on that used for the sirodesmin biosynthetic gene cluster. All genes within the cluster have a gli prefix. Genes with homologues in the sirodesmin biosynthetic gene cluster have the same suffix.
bFor genes outside the cluster, the LocusID from the Central Aspergillus data repository annotation has been added where the prediction presented here matched those at http://www.cadre.man.ac.uk/.
Homologies between the A. fumigatus putative gliotoxin biosynthetic gene cluster and the L. maculans sirodesmin biosynthetic gene cluster
| Gene namea | Sirodesmin cluster homologue | BLASTX match | e-value | Encoded product/commentsb |
| − | − | MG08409.4 | 7e−61 | Endoglucanase |
| − | − | AN6102.2 | 4e−84 | Quinone oxidoreductase |
| − | − | AN7527.2 | 5e−91 | Zinc finger transcription factor |
| − | − | AN7529.2 | 5e−150 | Peptidase (Afu6g09600) |
| − | − | CPPS2, Claviceps purpurea (CAD28788) | 2e−109 | Mono-modular peptide synthetase (Afu6g09610) |
| gliZ | sirZ | SirZ | 5e−10 | Zinc finger transcriptional regulator |
| gliI | sirI | SirI | 2e−61 | 1-aminocyclopropane-1-carboxylic acid synthase (ACCS). Core ETP biosynthesis. |
| gliJ | sirJ | MG07739.4 | 2e−92 | Dipeptidase. Core ETP biosynthesis. |
| gliP | sirP | SirP | 0 | Two module non-ribosomal peptide synthetase. Core ETP biosynthesis. |
| gliC | sirC | MG07743.4 | 3e−92 | Cytochrome P450 monooxygenase sharing similarity with SirC. Core ETP biosynthesis. |
| gliM | sirM | SirM | 1e−80 | O-Methyl transferase. Core ETP biosynthesis. |
| gliG | sirG | Alternaria alternata GST (AAR98813) | 3e−78 | Glutathione S-transferase. Core ETP biosynthesis. |
| gliK | none | MG07739.4 | 2e−35 | Hypothetical protein, no matches in sirodesmin biosynthetic gene cluster. |
| gliA | none | FG04188.1 | e−103 | Major facilitator type transporter. Functional equivalent of SirA. |
| gliN | sirN | SirN | 2e−75 | Methyl transferase. Core ETP biosynthesis. |
| gliF | none | AN1598.2 | 9e−54 | Cytochrome P450 monooxygenase. Not necessarily functional equivalents of SirB or SirE |
| gliT | sirT | AN3963.2 | 1e−70 | Thioredoxin reductase. Core ETP biosynthesis. |
| AN2610.2 | 0 | Cytochrome P450 monooxygenase trichothecene C15 hydroxylase (Tri11) (Afu6g09760) | ||
| − | AN2611.2 | e−117 | Domain matches to terpene cyclase and transisoprenyl pyrophosphate synthase. Probably involved in terpenoid (secondary) metabolism. (Afu6g09770) | |
| − | AN9390.2 | 3e−95 | Chitinase (Afu6g09780) |
| Gene namea | Sirodesmin cluster homologue | BLASTX match | e-value | Encoded product/commentsb |
| − | − | MG08409.4 | 7e−61 | Endoglucanase |
| − | − | AN6102.2 | 4e−84 | Quinone oxidoreductase |
| − | − | AN7527.2 | 5e−91 | Zinc finger transcription factor |
| − | − | AN7529.2 | 5e−150 | Peptidase (Afu6g09600) |
| − | − | CPPS2, Claviceps purpurea (CAD28788) | 2e−109 | Mono-modular peptide synthetase (Afu6g09610) |
| gliZ | sirZ | SirZ | 5e−10 | Zinc finger transcriptional regulator |
| gliI | sirI | SirI | 2e−61 | 1-aminocyclopropane-1-carboxylic acid synthase (ACCS). Core ETP biosynthesis. |
| gliJ | sirJ | MG07739.4 | 2e−92 | Dipeptidase. Core ETP biosynthesis. |
| gliP | sirP | SirP | 0 | Two module non-ribosomal peptide synthetase. Core ETP biosynthesis. |
| gliC | sirC | MG07743.4 | 3e−92 | Cytochrome P450 monooxygenase sharing similarity with SirC. Core ETP biosynthesis. |
| gliM | sirM | SirM | 1e−80 | O-Methyl transferase. Core ETP biosynthesis. |
| gliG | sirG | Alternaria alternata GST (AAR98813) | 3e−78 | Glutathione S-transferase. Core ETP biosynthesis. |
| gliK | none | MG07739.4 | 2e−35 | Hypothetical protein, no matches in sirodesmin biosynthetic gene cluster. |
| gliA | none | FG04188.1 | e−103 | Major facilitator type transporter. Functional equivalent of SirA. |
| gliN | sirN | SirN | 2e−75 | Methyl transferase. Core ETP biosynthesis. |
| gliF | none | AN1598.2 | 9e−54 | Cytochrome P450 monooxygenase. Not necessarily functional equivalents of SirB or SirE |
| gliT | sirT | AN3963.2 | 1e−70 | Thioredoxin reductase. Core ETP biosynthesis. |
| AN2610.2 | 0 | Cytochrome P450 monooxygenase trichothecene C15 hydroxylase (Tri11) (Afu6g09760) | ||
| − | AN2611.2 | e−117 | Domain matches to terpene cyclase and transisoprenyl pyrophosphate synthase. Probably involved in terpenoid (secondary) metabolism. (Afu6g09770) | |
| − | AN9390.2 | 3e−95 | Chitinase (Afu6g09780) |
Not all gli gene products had a best match to the equivalent from L. maculans. Some showed higher sequence similarity to products encoded by the M. grisea cluster [[2]. Four (GliA, GliF, GliG and GliT) showed higher sequence similarity to gene products from other species, but all these are members of multigene families.
aGene nomenclature is based on that used for the sirodesmin biosynthetic gene cluster. All genes within the cluster have a gli prefix. Genes with homologues in the sirodesmin biosynthetic gene cluster have the same suffix.
bFor genes outside the cluster, the LocusID from the Central Aspergillus data repository annotation has been added where the prediction presented here matched those at http://www.cadre.man.ac.uk/.
Despite the significant sequence similarity between the encoded products of the sirodesmin and gliotoxin biosynthetic gene clusters, the arrangement of the genes within the clusters differs. However, there are some similarities. The thioredoxin reductase genes (sirT and gliT) are at the end of each cluster. The transporter and sirN/gliN genes are also adjacent to each other, as are sirC/gliC and sirM/gliM, and sirI/gliI and sirJ/gliJ, although directions of transcription are altered, except for the transporters and sirN/gliN.
The remnant of a retroelement that appears to have undergone repeat induced point (RIP) mutation is immediately adjacent to gliT (Fig. 2). RIP mutation brings about the change of C to T at CA dinucleotides, resulting in an increase in AT content and under-representation of CA dinucleotides [[5]. The AT content of this region is 64% compared to 47% for the genic regions of the sequence represented in Fig. 2 and is lacking CA dinucleotides compared to surrounding areas. Distal to this is a pair of genes, which are conserved in A. nidulans (AN2610.2 and AN2611.2). These genes are also homologous to genes involved in secondary metabolism in other fungi (Table 2). At the other end of the cluster, adjacent to gliZ, there is a group of genes with a possible role in secondary metabolism. These include a homologue of the C. purpurea mono-modular peptide synthetase gene cpps2 involved in ergotamine biosynthesis [[6]. Homologues of the hypothetical proteins, AN7527.2 and AN7529.2, which in A. nidulans are interrupted by only one gene, may also have a role in secondary metabolism. A homologue of AN7524.2 is present in the region to the left of that shown in Fig. 2, suggesting that the gene order in this region is partially conserved between A. fumigatus and A. nidulans. However, these genes are interrupted by at least two other predicted genes that do not have linked homologues in A. nidulans. Also one of these (AN6102.2) appears to have been interrupted by a genetic element in A. fumigatus but not in A. nidulans (Fig. 2). This element also appears to have undergone RIP-like mutation.
3.2 Time course of gliotoxin production
RP-HPLC profiles of A. fumigatus culture filtrates showed a peak of identical retention time to that of a commercial sample of gliotoxin (Supplementary figure 2). The identity of this peak as gliotoxin was confirmed by GC–MS (Supplementary figure 3). However, while a compound with a mass spectrum identical to the authentic sample was observed, gliotoxin was converted to a related compound under the conditions used for GC–MS which has also been reported by others [[7,[8].
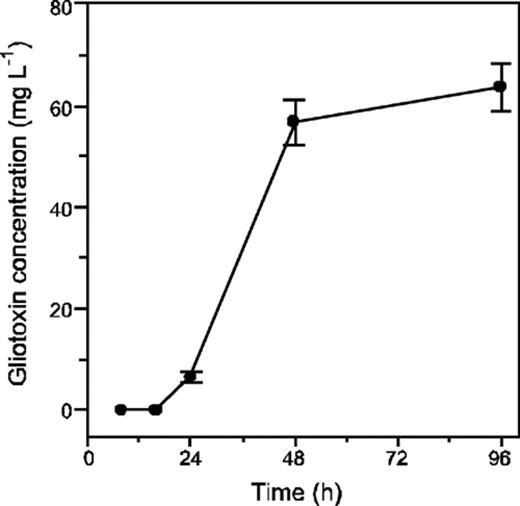
The production of gliotoxin over time was analysed by RP-HPLC. Gliotoxin was first detectable in culture filtrates after 24 h and increased most rapidly between 24 and 48 h (Fig. 3).
Gliotoxin production by Aspergillus fumigatus isolate Af293. Cultures were grown in Czapek Dox media at 37°C with agitation and gliotoxin was detected by RP-HPLC and quantified by comparison to a standard preparation. Values are expressed as the mean of two extractions from the same culture filtrate.
3.3 Genes in the cluster are co-regulated with gliotoxin production
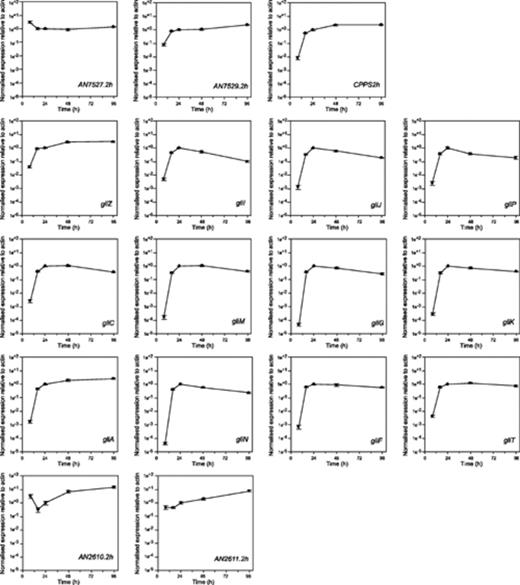
The expression of 12 genes predicted to be part of the gliotoxin biosynthetic gene cluster was coregulated (Fig. 4) and the timing of expression corresponded with the production of gliotoxin (Fig. 3). Transcripts for predicted biosynthetic enzymes all peaked in expression at either 24 or 48 h with the level of expression at both times being similar, which corresponds to the time at which the rate of gliotoxin biosynthesis was maximal. The gene encoding the transporter, gliA peaked at the end of the time course, when the level of gliotoxin in the media was maximal. The putative transcriptional regulator gene, gliZ, also peaked at the end of the time course. The pairwise correlation coefficient of logarithmically transformed data was in most cases greater than 0.9 for all “gli” genes. The correlation between the gli genes and AN7529.2 and cpps2 homologues was also greater then 0.9. However, while the data analysis method does not allow for absolute comparison between genes (as the efficiency of reverse transcription for different genes is not considered), the expression level of the cpps2 homologue was significantly lower (approximately three orders of magnitude) than the gli genes (data not shown). The other three genes tested that flank the putative cluster showed a different expression pattern to those in the cluster, confirming earlier predictions of the physical limits of the gene cluster (Fig. 4).
Co-regulation of genes in the gliotoxin biosynthetic gene cluster of Aspergillus fumigatus isolate Af293 and five flanking genes. The cultures used for RNA extraction were the same as those used for analysis of gliotoxin production (Fig. 3). Data are presented as gene expression relative to actin normalised to the level of gene expression at 24 h. The absolute expression level of the cpps2 homologue was significantly lower (approximately three orders of magnitude) than the gli genes.
4 Discussion
Bioinformatic analyses show that the genes within the A. fumigatus gene cluster and the sirodesmin biosynthetic gene cluster are very similar. Putative biochemical roles for the genes conserved between the two clusters are discussed elsewhere [[,[2]. The absence of the A. fumigatus cluster from the genome of the closely related, gliotoxin non-producing fungus, A. nidulans and the structural similarities between the gliotoxin and sirodesmin molecules suggests strongly that these A. fumigatus genes comprise the gliotoxin biosynthetic gene cluster. Based on an predicted similarity in the biosynthetic pathway, which probably does not include any extra modifications of the phenylalanyl residue compared to those of the tyrosyl residue in sirodesmin biosynthesis, additional genes were not expected in the biosynthetic gene cluster for gliotoxin production. However, two additional genes, gliF which encodes a cytochrome P450 monooxygenase and gliK, which encodes a protein with best match to a hypothetical protein were present. Cytochrome P450 monooxygenases typically introduce hydroxyl groups and it is possible the “extra” cytochrome P450 monooxygenase, GliF, in the gliotoxin gene cluster may add the hydroxyl group to the phenylalanine residue. However, this hydroxyl group is probably derived from an arene oxide intermediate (second bracketed compound in Fig. 1) and not from direct addition of a hydroxyl group [[9]. Similarly in sirodesmin biosynthesis a hydroxyl group is likely to be introduced via this type of intermediate [[2,[0]. The function of GliK in gliotoxin biosynthesis is also unknown and difficult to predict.
The remnant of a transposable element that appears to have undergone RIP-like mutation, adjacent to gliT is an interesting feature of the cluster. A repetitive element is also adjacent to LmHDX1 near the sirodesmin biosynthetic gene cluster of L. maculans (Cozijnsen and Howlett, unpublished data). RIP mutation, which occurs during pairing of homologous sequences during meiosis and results in C/G to A/T transitions usually at CA dinucleotides, would not be expected to occur in A. fumigatus as this fungus does not reproduce sexually [[5]. However, the events that led to the increase in AT content in this region may be ancestral. Whether the presence of transposable elements adjacent to these secondary metabolite gene clusters has any relevance to their evolution is unknown.
The presence of a cpps2 homologue in A. fumigatus linked to the putative gliotoxin cluster is interesting. In C. purpurea cpps2 encodes LPS2, an enzyme responsible for the incorporation of d-lysergic acid into ergopeptines [[6]. Such a gene would not be expected to be involved in gliotoxin biosynthesis. However, A. fumigatus makes a group of similar compounds to d-lysergic acid, namely fumigaclavines [[1]. Whether the fumigaclavines are incorporated into molecules similar to ergopeptines is unknown.
The correlation between the timing of expression of the genes in the A. fumigatus cluster and production of gliotoxin further supports the assertion that the cluster is responsible for gliotoxin production. The synthesis of gliotoxin by A. fumigatus largely occurred in a short period in the culture time course (i.e., between 24 and 48 h), although there was some continued production after this point, as is seen for gliotoxin production in Trichoderma virens[[2]. This experiment also provided evidence for the limits of the cluster as was done for the sirodesmin biosynthetic gene cluster [[2] and for other metabolite gene clusters [[3,[4]. Correspondence between gene expression and a phenotype of interest is powerful, albeit correlative, evidence for a link between gene and phenotype. Indeed, gene expression coregulation is the basis of defining new pathways and interactions in many microarray based experiments [[5,[7].
Gliotoxin production by A. fumigatus occurs earlier than sirodesmin production in L. maculans, which probably reflects the faster growth rate of A. fumigatus relative to L. maculans and also the different culture conditions used, particularly temperature [[2]. However, the trend of a large (at least two orders of magnitude) increase of gene expression at the times that the compounds were first detected in culture filtrates was similar for genes in both clusters. The similarities in expression pattern between the two clusters indicate that the pathways that regulate gene expression in response to nutrient status are probably similar, as both organisms were cultured in complete media. However, these experiments do not indicate potential similarities in regulatory networks involved in response to the host environment.
In summary the correlation of the timing of gene expression with the production of gliotoxin is strong evidence that the A. fumigatus gene cluster is responsible for gliotoxin production. Additionally the high level of sequence similarity between the L. maculans and A. fumigatus gene clusters, and lack of similar clusters in A. nidulans further supports this contention. If genes in this cluster are mutated and the resultant mutants are unable to produce gliotoxin, this will unequivocally prove that the cluster described here encodes enzymes in the gliotoxin biosynthetic pathway.
Acknowledgements
We thank Professor Geoffrey Turner, The University of Sheffield for providing A. fumigatus isolate Af293. Preliminary sequence data for A. fumigatus were obtained from The Institute for Genomic Research website at http://www.tigr.org. Sequencing of A. fumigatus was funded by the National Institute of Allergy and Infectious Disease U01 AI 48830 to David Denning and William Nierman, the Wellcome Trust, and Fondo de Investicagiones Sanitarias.
References